Antibodies targeted to histone modifications may bind non-specifically to similar, but off-target histone modifications. Conversely, their specific binding can be inhibited by steric hindrance from modifications on neighboring residues. Assays like ELISA, western blot, ChIP, and IF are commonly used to demonstrate antibody specificity and sensitivity, but they cannot clearly predict how an antibody will interact with nearby epitopes. As a result, an alternative approach is needed when trying to validate an antibody to a histone modification target. Continue reading to see how we do it.
CST modification-specific histone antibodies are validated using a peptide array assay similar to the one described by Fuchs, S.M., et al. (1). In a single experiment, these arrays assess reactivity against known modifications across all histone proteins as well as the effects of neighboring modifications on the ability of the antibody to detect a single modification site. Therefore, the peptide array assay allows us to confirm the antibodies are performing as expected.
Step 1: Array
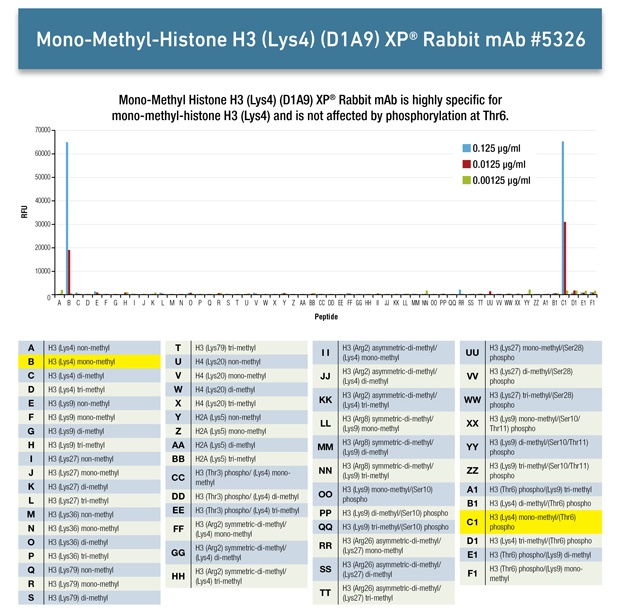
Peptides with mono-, di-, tri-methyl, acetyl-, or unmodified lysine are spotted onto nitrocellulose either alone or in combination with a known neighboring histone modification (e.g., histone H3K4Me3), as indicated in the diagram. We use a similar array to test our methyl-arginine antibodies.
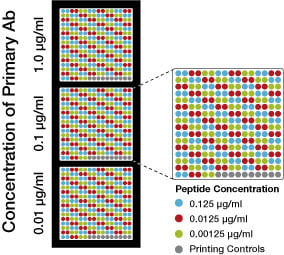
Step 2: Antibody
The histone modification antibody is then applied to the array at at three concentrations, as indicated in the diagram. This allows us to assess antibody reactivity while ensuring that the concentration is not saturating the assay.
Step 3: Analysis
The arrays are washed and incubated in a fluorescently tagged secondary antibody and then read using a LI-COR® Odyssey® Infrared Imager.
Step 4: Data
Here is the analysis we performed for Mono-Methyl-Histone H3 (Lys4) (D1A9) XP® Rabbit mAb #5326
See more examples of validation of histone modification antibodies.
If you'd like to see how we stack up against other Histone Modification Antibodies, you can click/tap below to download a performance comparison brochure, or check out this interactive database (Rothbart, et al., 2015) that compares peptide array data for widely-used histone modification antibodies from commercial sources.
(1) Fuchs, S.M., et al. (2011) Curr. Biol. 21, 53-58.
LI-COR and Odyssey are registered trademarks of LI-COR Biosciences.