Since the discovery of antibodies, decades of research and development have spurred technological advances, leading us to where we are today—able to readily purchase antibodies as off-the-shelf reagents for scientific research. In Part 2 of the Antibody Essentials series, we discussed how antibody diversity is generated, and how antibodies are classified. Here, in Part 3, we'll cover how antibody technology was first discovered and how it has evolved over the past decades.
Understanding the background and historical development of antibody-based technologies will help you to appreciate the uses and limitations of antibodies in modern experimental techniques. Even though it's easier than ever before to buy antibodies for your research, we encourage you to think about antibodies as if you were producing them yourself. This will greatly increase your chances of generating results you can trust.
Table of Contents
- Antibodies: The Origin Story
- Leveraging Antibodies as Research Tools
- Acquiring Antibodies: Then vs Now
- How are Peer Review and Reagent Validation Connected?
- The Modern RUO Antibody Marketplace
- Questions to Ask When Selecting Antibodies for Your Experiment
Antibodies: The Origin Story
The road leading to the discovery of antibodies and their use as essential research reagents began in the early 18th century with the first smallpox inoculation studies. By transferring material from the sores of human smallpox patients into scratches made in the skin of healthy individuals, it was shown to be possible to protect against future smallpox infections. Importantly, these experiments provided some of the very earliest insights into the mechanisms underlying immunity. Due to safety concerns, the method was adapted by Edward Jenner in 1796 to use material taken from the sores of individuals with cowpox—a much milder disease than smallpox. Louis Pasteur then introduced further improvements in 1885 when he used attenuated viruses to provide protection without risking the development of disease.
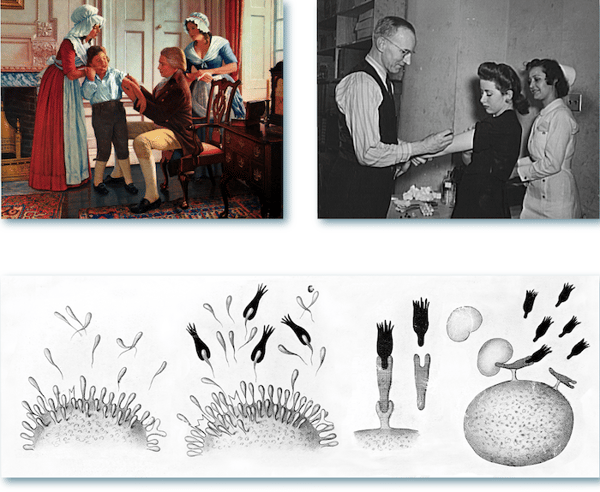 Figure 1. Top left: Edward Jenner, English rural physician, performed the first vaccination against smallpox at Berkeley in 1796. (Source: US Nat’l Library of Medicine.) Top right: US Office of War Information employees receiving free inoculation against smallpox, diphtheria, and typhoid. (Source: US Library of Congress.) Bottom: Paul Ehrlich's side-chain theory laid the conceptual groundwork for the discovery of antibodies. (Source: Wellcome Collection.)
Figure 1. Top left: Edward Jenner, English rural physician, performed the first vaccination against smallpox at Berkeley in 1796. (Source: US Nat’l Library of Medicine.) Top right: US Office of War Information employees receiving free inoculation against smallpox, diphtheria, and typhoid. (Source: US Library of Congress.) Bottom: Paul Ehrlich's side-chain theory laid the conceptual groundwork for the discovery of antibodies. (Source: Wellcome Collection.)
After this, efforts to understand and harness the immune response advanced relatively quickly. In the late 19th century, a Nobel Prize was awarded to von Behring and Kitasatu for the discovery that serum taken from rabbits infected with tetanus or diphtheria could confer disease resistance to mice. This finding was followed by Ehrlich’s side-chain theory (Figure 1, bottom), where he hypothesized that immune cells are covered with receptors able to bind specifically to a foreign substance. According to Ehrlich, once such an interaction had taken place, the immune cells would become activated to produce more receptors, which would then be released into the bloodstream to help eliminate the unwanted agent.
The side-chain theory is widely recognized as the first description of what we now know to be antibodies. After this, an explanation of the antibody-antigen binding hypothesis was suggested, with B cells being identified as the source of antibodies shortly thereafter.
 |
Blog: Do You Trust Your Research Antibody? |
Leveraging Antibodies as Research Tools
The latter half of the 20th century saw the determination of the molecular structure of antibodies and the invention of hybridoma technology, in which an immune response is evoked in vivo to expand and select B cell clones (covered in detail in Part 4 of Antibody Essentials)—providing Nobel prizes for Edelman and Porter, and Jerne, Köhler, and Milstein, respectively.
 Figure 2. Top row: Gerald M. Edelman & Rodney R. Porter; Bottom row: Niels K. Jerne, César Milstein, & Georges J.F. Köhler. (Source: US Nat’L Library of Medicine (top and lower left); MRC Laboratory of Molecular Biology (lower right).
Figure 2. Top row: Gerald M. Edelman & Rodney R. Porter; Bottom row: Niels K. Jerne, César Milstein, & Georges J.F. Köhler. (Source: US Nat’L Library of Medicine (top and lower left); MRC Laboratory of Molecular Biology (lower right).
Many immunoassay techniques were also invented around that time, including flow cytometry, Enzyme-linked immunosorbent assay (ELISA), western blot, and chromatin immunoprecipitation (ChIP). With the development of polymerase chain reaction (PCR) and associated modern molecular cloning techniques, hybridoma technology has been complemented by various recombinant methods for generating antibodies, greatly expanding the diversity of antibody tools available. Another notable development was the publication of Harlow and Lane's book, Antibodies: A Laboratory Manual. Following its release in 1988, this book was soon found on the shelves of labs and science libraries worldwide, rapidly becoming a go-to reference for scientists who wanted to develop antibodies for their research.
In the 90s, technical advances also led to the development of antibodies that could specifically detect post-translational modifications (PTMs), starting with phospho-specific antibodies and followed by tools to detect methylation, acetylation, and other protein modifications. This was important because such modifications are associated with changes in protein activation states and biological function, such as turning signaling pathways on or off, meaning these antibody reagents opened up new and exciting possibilities for characterizing signaling activities. This was also the period when Cell Signaling Technology (CST) was founded.
Gradually, the number of research antibodies available to purchase, from CST and other vendors, began to rise, setting the stage for the exponential growth of commercial antibodies seen in the past decade. Today, you might have many options when it's time to buy antibodies for your experiment, depending, of course, on your chosen target and application. Publications citing the use of research use only (RUO) antibodies have grown steadily in number year after year.
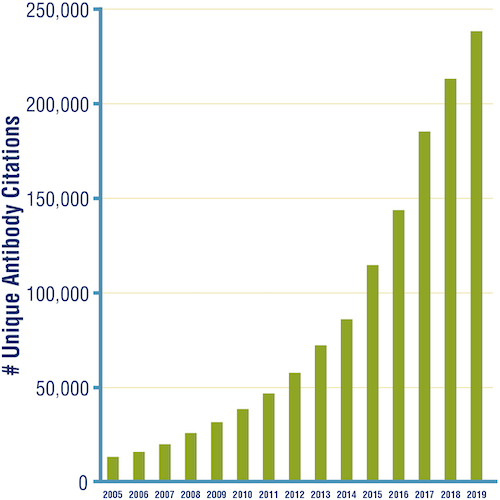 Figure 4. Number of unique citations in publications for primary antibodies from all vendors, 2005–2019. (Source: CiteAb.)
Figure 4. Number of unique citations in publications for primary antibodies from all vendors, 2005–2019. (Source: CiteAb.)
Acquiring Antibodies: Then vs Now
With easy access to so many antibodies, and research publications readily publishing the results of those antibodies, why has the reproducibility crisis emerged? Part of the reason may be that it is now almost too easy to buy an antibody in a way that wasn't possible before. To help you understand how the research landscape has changed with respect to antibodies, let's take a trip back in time to the 80s or 90s, when hybridoma and molecular cloning technologies were available, but before many antibodies were commercially available.
Let's imagine that you're a scientist in this time period, working in a university lab, and you’re studying a novel target protein that isn’t yet well understood. You’re writing a research fellowship grant and you’re hoping to generate new publications. Obviously, you couldn’t buy an antibody to a protein that’s just been discovered, so as one of the research aims, you propose to develop a polyclonal or monoclonal antibody to use in your experiments. Your research plans might involve using the new antibody in several different applications, modifying existing experimental protocols, or even designing new assays.
Figure 5. A research lab in the 1980s, before the widespread availability of commercial RUO antibodies. (Source: US Nat’l Library of Medicine.)
Keep in mind that in past decades, there would be less overall knowledge in the literature about your target protein that you could use to design validation tests. There might be less known about related proteins that might share functions, structure, or sequence homology, so you'd want to be cognizant of this possibility when developing and validating a novel antibody that might react with your protein plus an unknown protein. You would also want to stay current on recent literature to incorporate new information, for example details about subcellular localization, post-translational modifications, or binding interactions involving your protein, when validating your antibody. Scientific knowledge builds on itself, and some of that knowledge informs antibody development and validation.
%20sc-7480%20vs%20CST%20Bax%20Antibody.webp?width=250&height=202&name=Santa%20Cruz%20(B-9)%20sc-7480%20vs%20CST%20Bax%20Antibody.webp) |
Blog: Watch Your Bax: How a Faulty Antibody May Have Cost Millions in Flawed Research |
How are peer review and reagent validation connected?
One aspect of academic research from earlier decades that remains true today is the dual and interdependent needs to get funding and to publish research. Throughout the processes of grant writing, research execution, and manuscript publishing, you’re always anticipating critical comments from peer reviewers. (And from that one professor who always sits in the front row during seminar and asks the most cutting questions. Every department has at least one of those.) This motivates the design of better experiments that can stand up to peer review and support stronger conclusions.
Since robust experimental design applies to all aspects of a study, in our 1980s scenario, you can be sure the peer reviewers would want to see data to support that your antibody is specific and sensitive in the applications and assays that you use it in. In other words, you'd need to spend more time generating validation data before you could publish. To go from immunization to a polyclonal antibody takes months, developing a monoclonal antibody can take up to or over a year, and validation testing can add yet more time. In the 1980s, you would be both designing and executing your own antibody validation testing, and optimizing your protocols. It’s a safe bet that it would be something you’d spend a lot of time thinking about. All of this lead time prior to experimental scale-up and data collection and analysis would be worth it, however, because you would be able to trust your results and be confident when it comes time to publish.
The Modern RUO Antibody Marketplace
Now, let’s fast forward to today. There are still research labs that choose to make and validate their own antibodies, such as labs that study somewhat specialized research areas or use less-common model organism species. But it’s much more likely that your first instinct will be to read the literature and type search terms into Google or Baidu or go to a reagent-focused shopping engine such as Biocompare, CiteAb, or Antibody Resource. This allows you to compare antibodies before choosing one to buy and have shipped to your lab. Today, antibody end-users (i.e., researchers) are aware of the reproducibility crisis that we introduced in the first Antibody Essentials blog. During the reagent selection and shopping process, researchers will evaluate the validation data supplied by the antibody manufacturer and compare it with data generated by other scientists using that antibody that has been published in research journals.
Nevertheless, it’s a shifted perspective—instead of waking up in the middle of the night worrying about how to validate your antibody, you might consider validation to be the antibody vendor’s responsibility. And let’s be honest here, you may or may not have the same critical eye when looking at a vendor’s validation data as when you’re designing and executing the validation tests yourself with the expectation that they’ll be dissected and analyzed in detail by peer reviewers. It‘s also the case that research papers have become longer and denser, meaning antibody validation data tends to get stuffed in the supplemental methods sections, where it's easy to be missed, or it may just be omitted entirely. Overall, the reduced scrutiny of antibody validation and unintentional misuse of antibodies outside of their intended applications or biological models, even including antibodies with many research citations, have contributed to the reproducibility crisis.
When you’re searching on Google and browsing antibody vendor websites, you may find there’s a great deal of variability in what information is available when it comes to validation. You might see multiple vendors selling antibodies to the same target, but the validation for some of those antibodies does not support specificity and sensitivity in the desired application. Or, critical details might be missing from the validation data, such as what cell lines were used, how treatments were applied, or exactly what antibody dilutions were made. These shortcomings can make it difficult to ascertain the strength of validation data, potentially leading to misinterpreted experiments and the publication of irreproducible research in the literature.
Many vendors perform the validation testing themselves, which is standard practice for CST antibodies. But you may not be aware that some other antibodies are re-sold. In other words, the vendor did not produce and validate the antibody in their facility, but instead bought the antibody from a third party and put their label on the tube. Many of these re-branded antibodies can work well in their intended application, but it's a potential source of confusion when you think you are comparing two different antibodies to the same target, but they are in fact the same antibody being sold by two different brands.
Another source of variability may be different standards for lot-to-lot validation; some vendors will re-validate each new lot of antibody for each of its recommended applications, while others may not (at CST, we validate every antibody lot). As we shall see in the next Antibody Essentials blog, the composition of polyclonal antibody products can vary over time, so lot-specific validation is especially important for these reagents.
And finally, it’s worth noting that in some regions of the globe, there have been reports of nefarious vendors and resellers who dilute products or even sell counterfeit antibodies or research reagents. Fortunately, governments, vendors, and the scientific community are taking steps to combat these activities.
Questions to Ask When Selecting Antibodies for Your Experiment
We’d like to recommend that when you buy a commercially available antibody—or when you “borrow an aliquot” from your friend in the lab down the hall—you should think about it the same way you would if you were making and validating the antibody in your own lab. Put yourself in the shoes of a scientist in the 80s or 90s, who didn’t have their choice of several commercially developed antibodies, each linked to several publications.
Ask yourself: if you were making it, would you be confident submitting the “validation data” as a figure knowing it would get the attention of peer reviewers? Does the protocol used to validate the antibody closely align with your experimental protocols? Are the tissues or cells used in the vendor’s validation suitably informative in the context of what’s known about your experimental model? If you're assessing research papers that used the antibody you're evaluating, did the authors provide enough detail to confirm they used it "following the manufacturer's protocol", and did they perform additional validation when necessary?
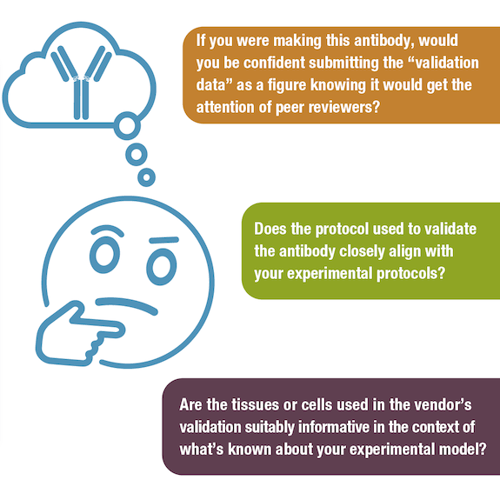
If you answered “no” to any of those questions, you may decide that it’s still worth setting up pilot experiments with the antibody in question. However, you'll want to supplement the vendor’s validation data with your own validation testing relevant to your chosen application and model system. Only after re-validating the antibody and optimizing your protocol (when necessary) should you proceed with scale-up and data collection. Keeping a detailed lab notebook will help when it comes time to write the methods section and publishing, so that peer reviewers and readers will be able to understand exactly how your results were generated.
So, once again, there’s a shared responsibility between the vendor (including CST), the antibody users (including you), scientific mentors, and publications to ensure antibodies are properly validated, used, and documented to improve reproducibility. By staying mindful of how antibodies are produced and validated, you can give yourself a better chance of generating reliable and reproducible experimental results.
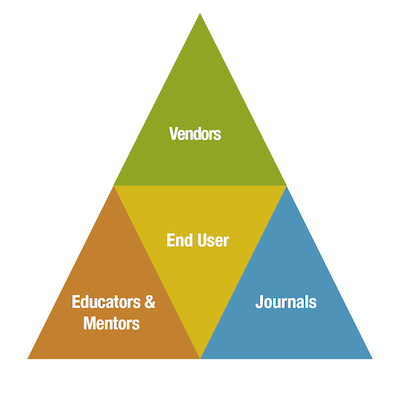 Figure 7. Scientific reproducibility, which includes the responsible use of antibodies in research, requires the continued cooperation of the scientific community as a whole.
Figure 7. Scientific reproducibility, which includes the responsible use of antibodies in research, requires the continued cooperation of the scientific community as a whole.
Hopefully, this overview of the discovery of antibodies and the evolutionary process bringing us to where we are today has provided an appreciation of the importance of antibodies to accelerating scientific research. Next, we’ll look in more detail at polyclonal and monoclonal antibodies, before comparing the advantages and disadvantages of antibodies from rabbit and mouse hosts.
Next up: Antibody Essentials Part 4: Polyclonal vs. Monoclonal Antibodies.
Read the entire Antibody Essentials series:
- Antibody Essentials Part 1: Antibody Basics
- Antibody Essentials Part 2: Antibody Diversity and Classification
- Antibody Essentials Part 3: How Antibody Technologies Evolved
- Antibody Essentials Part 4: Polyclonal vs Monoclonal Antibodies
- Antibody Essentials Part 5: Important Considerations for Antibody Selection
20-FLE-62535







