As a scientist, you probably already appreciate the vital role of antibodies in research. You may also be aware that the research community has been talking a lot about antibody validation over the past few years. The aim of the Antibody Essentials series is to clear up some of the confusion around what makes a good antibody and to provide guidance on the types of questions you should be asking when you’re choosing antibodies to support your research. Being an educated consumer and taking the time to understand the properties and limitations of different antibodies will help you to generate reliable immunoassay data and publish your findings with more confidence.
Here in Part 1, we’ll touch on the research reproducibility crisis and the importance of antibody validation, before running through some antibody basics including structure and function, binding properties, and the types of assays in which antibodies might be used. Later, we’ll introduce the concept of antibody validation, and why different antibody properties are so important.
Table of Contents
The Antibody Reproducibility Crisis
What do people mean when they talk about the research reproducibility crisis? In recent years, it’s become increasingly apparent that many experimental findings in the literature simply can’t be reproduced.1-4 You may have even been there yourself—trying to repeat an experiment that another lab published without success. Or, one day your experiment is working perfectly, and the next day it stops, and you have to figure out why. There are many potential sources of reproducibility issues:
- Experimental design
- Flawed analysis or data interpretation
- Contamination of cell lines
- The complexity of model organisms
- Reagents that have been poorly characterized or misused
Antibodies fall into the last category, and while they are not solely to blame for the reproducibility problem, they can certainly play a role.

Figure 1. During the 2010s, the research community began to question why many studies could not be replicated, a phenomenon called the Reproducibility Crisis.
Broadly speaking, several factors may contribute to a lack of experimental reproducibility with antibodies. First, the company that supplied the antibodies may have performed little, if any, quality control in the form of validation testing.
Second, the end-user (meaning the researcher who purchased the antibody) may not have considered the differences between their experimental model and the models used by the vendor to validate the antibody, instead choosing to trust that the antibody will perform as advertised without optimization. Furthermore, there may be differences in experimental protocols or conditions in the end-user's experiment compared to what’s been previously published. Unfortunately, insufficient detail in materials and methods of published research papers has been a contributor to the reproducibility problem, although the scientific and publishing communities have made strides to address this through peer and editorial review.
 |
Blog: Do You Trust Your Research Antibody? |
Finally, training and mentorship at the bench have a role to play. A lack of thorough training, combined with the pressure to publish, could certainly exacerbate the aforementioned issues. Improving reproducibility is a shared responsibility of the vendor, end-user, educators and mentors, and journals. As you go through your career as a research scientist, understanding how antibodies are made and how they work will greatly increase your chances of generating robust, reproducible data.
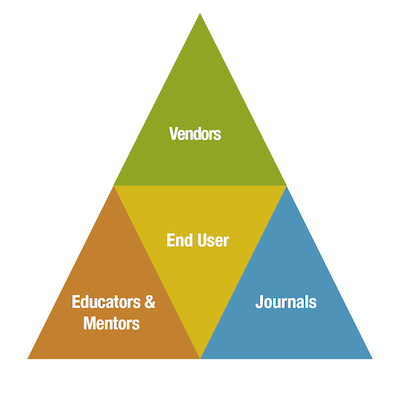
Figure 2. Reproducibility is a shared responsibility. Antibody vendors, end-users/researchers, mentors, and journals all have a role to play in improving the reproducibility of research.
What are antibodies, and how are they used in research?
Let’s start at the beginning: What, biologically speaking, is an antibody, and what makes an antibody useful as a tool in the lab? An antibody is a complex protein made by a particular class of immune cells called B cells during the adaptive immune response. The adaptive immune system, which originated in jawed fishes and is present in all higher vertebrates, confers a form of molecular memory that is both highly specific and incredibly diverse in its ability to recognize molecules from foreign sources, including pathogens. The human adaptive immune system has the theoretical repertoire to generate billions of unique antibodies as a result of the DNA rearrangement that occurs during B-cell development, which we will cover in an upcoming blog.
Since different antibodies should specifically recognize different epitopes and antigens, scientists can exploit antibody specificity to detect biomolecules in a research experiment, or in diagnostic applications such as disease biomarkers or pregnancy testing, or in therapeutic interventions such as cancer treatment.

Figure 3. Antibodies can be employed in many different types of application formats, or immunoassays.
When scientists talk about using antibodies for research, they’re most likely running some form of immunoassay. This term encompasses a wide range of techniques that use antibodies to detect, capture or neutralize a target. Common types of immunoassays or "applications" are shown above in Figure 3, and include western blot, immunoprecipitation, immunohistochemistry, immunofluorescence, mass spectrometry, among others.
Depending on the immunoassay that is chosen, a single antibody, pair of antibodies, or a complex panel of antibodies may be used. Mouse and rabbit antibodies are most commonly chosen for scientific research, although antibodies from rodents like rats and guinea pigs, and from hoofed mammals such as goats, sheep, and donkeys are also popular. In recent years, chicken antibodies and single-chain camelid antibodies have likewise become more mainstream. Whatever the immunoassay or antibody host species, the success of any immunoassay experiment hinges on the antibody having been validated and the protocol optimized for the intended application and experimental model. Otherwise, the data produced could lead to erroneous conclusions.
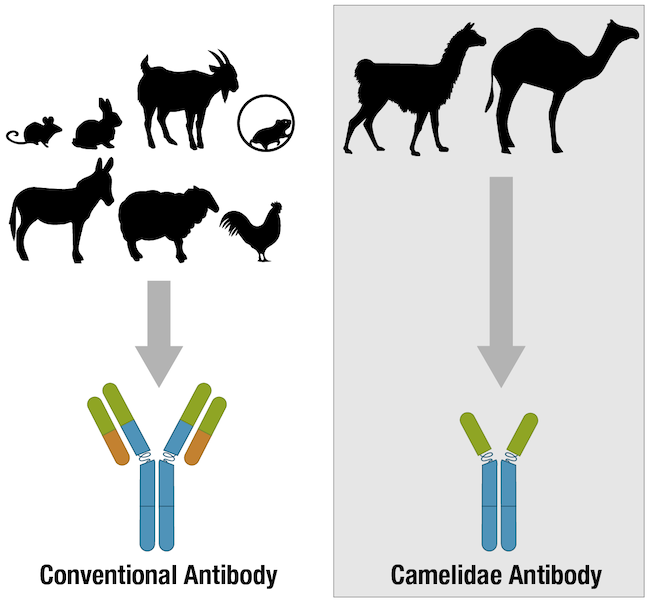
Figure 4. Many vertebrate species have antibodies, but most of the antibodies used for research are produced in mammalian species.
We should note that the term “validation” itself may mean slightly different things to different scientists. If you are in an academic lab, you’ll likely think of validation as it applies to a single antibody or reagent that is being used in an experiment. If you go on to work in a pharmaceutical or biotech company that does preclinical, translational, or clinical diagnostic work with patient samples, you may see the term “verification” instead of “validation” when evaluating the performance of an individual reagent. In the clinical setting, “validation” tends to be applied to a workflow (such as immunohistochemistry of biopsy material from patients for diagnostic purposes) as a whole, which includes the protocols and analysis, as well as reagents.
Going forward, we will use “validation” in the more academic definition, where we’re talking about validating the performance of an antibody rather than the whole experiment or workflow.
Antibody Validation at CST
At CST, we approach antibody validation with rigor and adhere to the Hallmarks of Antibody Validation. Essentially, this means that we validate all of our products for use in specific immunoassays. We recognize that just because an antibody has proven sensitive and specific for use in a western blot, for example, does not mean it will necessarily perform the same in a different assay using a different workflow. When searching for products on our website, you'll notice that the application(s) in which we have validated the antibody are listed for every product.
Keep reading, we'll talk more about exactly what it means for an antibody to be both sensitive and specific later on in this post, and in subsequent posts in this series.
Antibody Structure & Function
Generally, an antibody unit consists of four polypeptides: two identical heavy chains and two identical light chains. Each heavy chain contains three constant domains and one variable domain, whereas each light chain contains one constant domain and one variable domain. Intramolecular disulfide bonds stabilize domains, and intermolecular disulfide bonds hold the four chains together. Together, they form the characteristic Y shape you’re familiar with from textbooks.
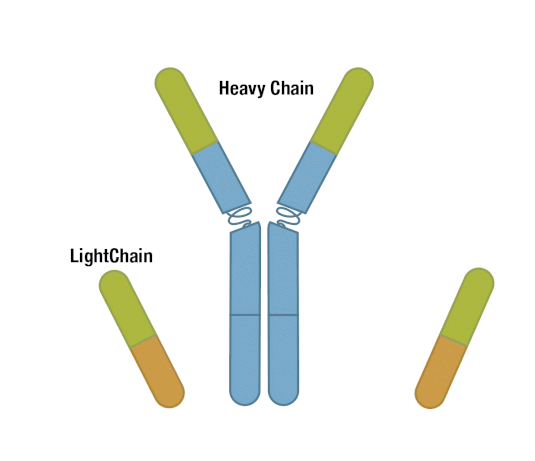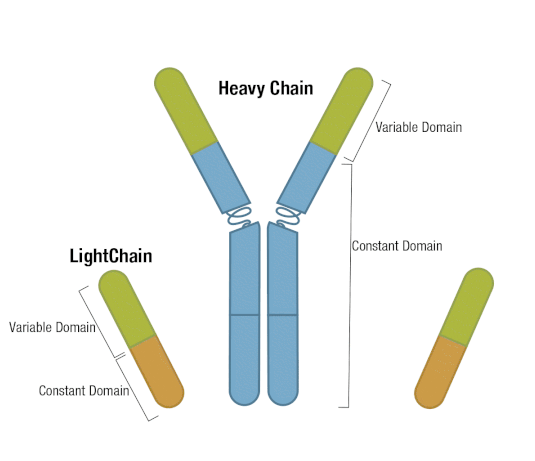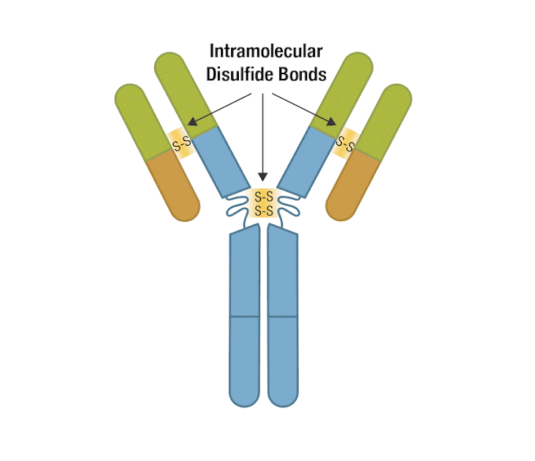
Figure 5. Each antibody macromolecule is composed of two heavy-chain and two light-chain polypeptides, held together by disulfide bonds.
The presence of proteolytic cleavage sites in the antibody enables digestion with enzymes such as pepsin or papain, resulting in antibody fragments with different characteristics. The effector end of the antibody is referred to as the Fc fragment. This interacts with Fc receptors on various immune cells during an immune response. The protein sequences of the Fc fragment, derived from the heavy chain, are highly conserved across all antibodies of the same isotype. The antigen-binding end of the antibody is known as the Fab fragment and, as you might expect, it plays a vital role in antigen recognition. The Fc fragment is separated from the Fab fragment by a hinge domain in the heavy chain, close to the location of the cleavage sites.
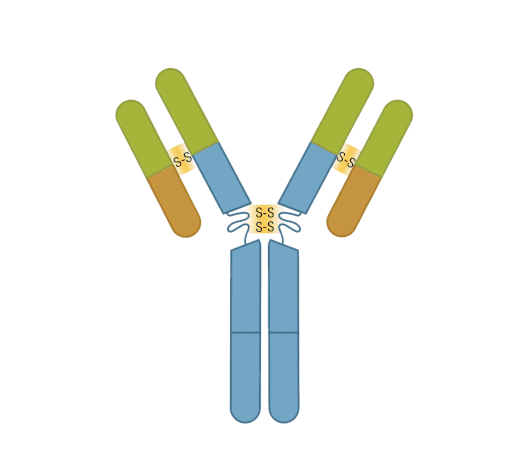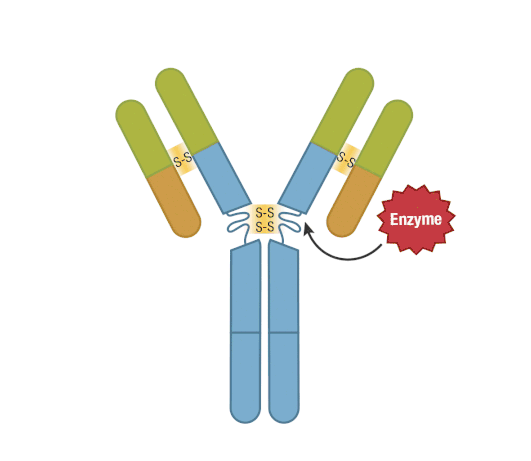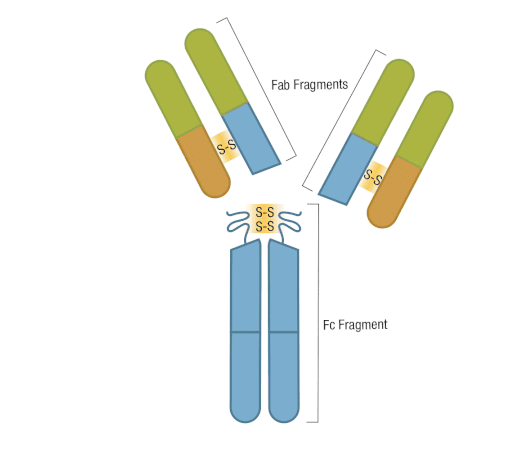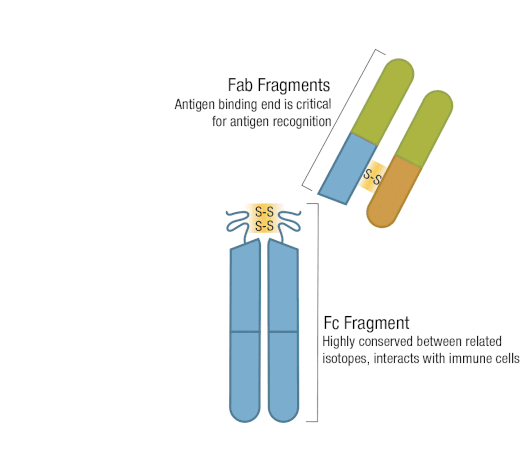
Figure 6. Enzymatic fragmentation can be used to create antibody fragments.
When an antibody encounters its antigen, a non-covalent, reversible binding event takes place. This may be mediated by a combination of hydrogen bonds, hydrophobic and electrostatic interactions, and van der Waals forces. The overall strength of the bond between the antibody and the antigen is defined by two key properties—affinity and avidity.
Antibody Affinity
First, let’s take a look at affinity. This describes the strength of the interaction between the antigen-binding site and the epitope and is usually measured by the equilibrium association constant, Ka. Ka is defined as the amount of antibody-antigen complex that exists at equilibrium or, putting it more simply, describes how quickly an antibody will bind to its antigen and for how long the two will stay bound. A high-affinity antibody with a high Ka will bind a greater amount of antigen in a shorter period of time than a low-affinity antibody with a low Ka.
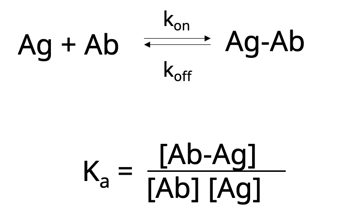
Now let’s take this one step further. Another way to think about affinity is the dissociation constant, Kd. This is simply the rate at which the antibody-antigen complex comes apart.
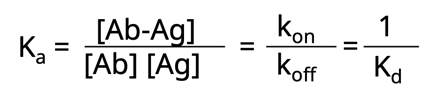 Both Ka and Kd are heavily influenced by numerous factors such as:
Both Ka and Kd are heavily influenced by numerous factors such as:
- Reaction size
- Buffer composition
- pH and temperature
When following or optimizing a protocol, it’s important to keep in mind that each of these variables can impact your experimental results.
Antibody Avidity
Next, let’s consider avidity, which is where things get a bit more complicated. Avidity describes the overall strength of the antibody-antigen complex. It is influenced not only by the number of available antigen-binding sites—better known as the antibody valency—but also by the affinity of each of those binding sites for the antigen. To put valency into context, think back to when we spoke about the very simplest antibody unit, a structure consisting of two heavy chains and two light chains, with two identical antigen-binding sites. This is a bivalent antibody. Most research antibodies are bivalent. However, naturally occurring antibodies with more than two antigen-binding sites exist, for example, IgA and IgM, and it’s also possible to engineer bispecific antibodies able to detect two distinct epitopes on the same target. While the antigen-binding sites in IgG antibodies tend to have a higher affinity for their antigens than the antigen-binding sites of IgM, the fact that IgM molecules are pentameric means they have ten binding sites, compared to two. This improves the avidity of IgM antibodies.
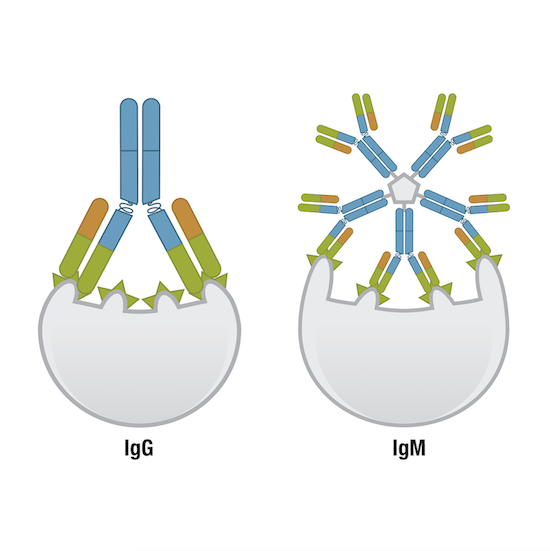 Figure 7. One IgG molecule has two antigen-binding sites, while one IgM molecule has ten, and thus, higher avidity than other immunoglobulin molecules.
Figure 7. One IgG molecule has two antigen-binding sites, while one IgM molecule has ten, and thus, higher avidity than other immunoglobulin molecules.
It’s also worth mentioning here that polyclonal antibodies typically have greater avidity than monoclonal antibodies because multiple polyclonals can bind a single target. In addition, the presentation of the antigen may be influenced by the sample processing methods that are used in different immunoassays. These may increase or decrease antigen availability or change the structure of the epitope, which can then affect avidity.
Affinity and avidity are not the only properties to consider when selecting and using an antibody. You should also consider the antibody specificity, selectivity & sensitivity.
Antibody Specificity, Selectivity, & Sensitivity
While many scientists are at least aware of antibody specificity as it relates to experimental reproducibility, some may not understand that antibody selectivity is an important concept as well. So, let's define and distinguish the two terms. The first of these is specificity, by which we mean the ability of the antibody to discriminate between its epitope, or region of a protein that determines antigenicity, from another. The second is selectivity, which describes how well an antibody binds its intended target molecule within a complex mixture.
When we consider specificity, it may be tempting to assume that an antibody with a high affinity and high avidity for its particular antigen would also be highly specific. In fact, this is not necessarily the case. While specificity is defined by the ability of the antibody to recognize and bind its intended epitope, it is independent of the strength of the antibody-antigen interaction. To better explain this, let’s consider a typical workflow to produce antibodies for research use, whereby animals are immunized with an antigen of interest. The resulting antibody pool will contain both high-affinity and low-affinity antibodies with similar specificities, yet the immune system generally favors expansion of the high-affinity antibody-producing B-cells. When the B-cells are isolated from the immunized animal and screened to identify those cells producing antibodies suitable for use as research reagents, a stringent selection process is necessary to ensure both high-affinity and low-affinity antibodies can be isolated. In this way, highly specific yet low-affinity antibodies won’t be overlooked.
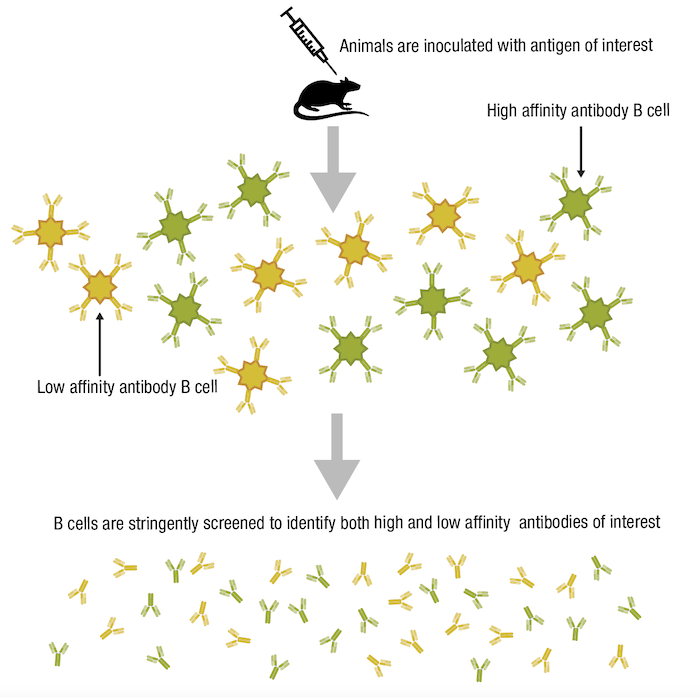
Figure 8. Lower-affinity antibodies may still be useful tools if they are highly specific.
Now let’s turn to selectivity, remembering that this describes how well an antibody binds to its intended target within a complex mixture. Although a highly specific antibody will detect its antigen in a given application and experimental context, all antibodies have the potential to bind to other biomolecules that may be present in a sample. Only some antibodies will be both specific for the desired epitope, and selective for the desired full-length target protein, as shown below in the scenario for a hypothetical Protein 1 in Figure 9.
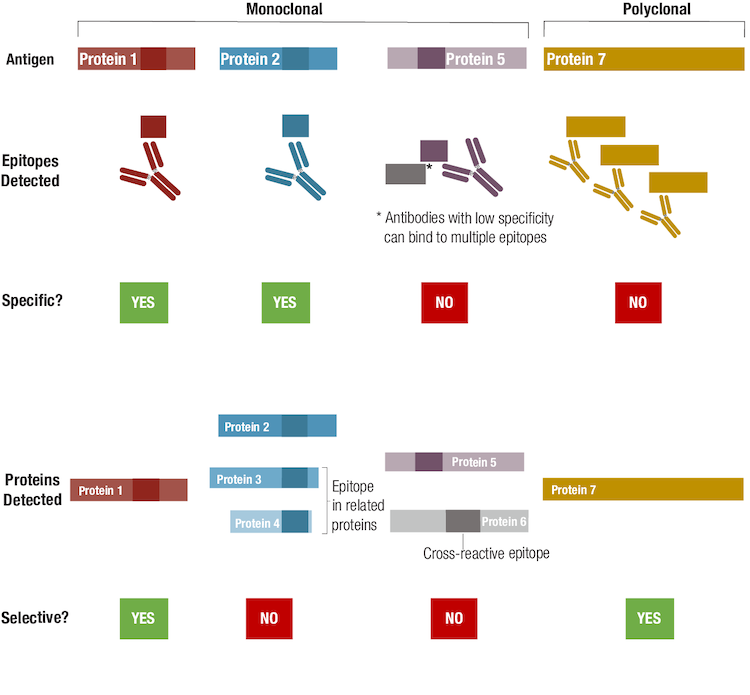 Figure 9. To understand the difference between antibody specificity and antibody selectivity, focus on the epitope(s) detected and protein(s) detected, respectively.
Figure 9. To understand the difference between antibody specificity and antibody selectivity, focus on the epitope(s) detected and protein(s) detected, respectively.
Even though an antibody could be specific for a single epitope, that epitope sequence could be present in other related, or even unrelated, proteins, and may contribute to the antibody having low selectivity. A hypothetical example for such a case is illustrated by the antibody generated for Protein 2, since the epitope recognized by this antibody is also expressed in related proteins. Note that while many researchers might consider this antibody to be non-specific, technically speaking it's more accurate to refer to it as "non-selective". Non-selective but specific antibodies may have utility as a "pan-reactive" tool, provided that interpretation of results accounts for the possibility of multiple isoforms or protein targets being detected. If an antibody detects the desired epitope but is also cross-reactive for one or more other epitopes, the antibody is neither specific nor selective, as shown in the example for Protein 5. The extent of this off-target binding, or cross-reactivity, can vary according to the experimental conditions and the relative abundance of the target, meaning that a poorly selective antibody can contribute significantly to unwanted background signal during an experiment.
%20sc-7480%20vs%20CST%20Bax%20Antibody.webp?width=250&height=202&name=Santa%20Cruz%20(B-9)%20sc-7480%20vs%20CST%20Bax%20Antibody.webp) |
Blog: Watch Your Bax: How a Faulty Antibody May Have Cost Millions in Flawed Research |
Finally, we consider a polyclonal antibody that detects multiple epitopes within Protein 7. Strictly speaking, the polyclonal is not specific in that it recognizes more than a single epitope. However, it can be said to be selective because only the intended antigen is detected by this polyclonal.
How do the properties of antibodies affect your experiments?
So now that we’ve covered affinity, avidity, specificity, and selectivity, what do these actually mean for your experiment? Although each of these properties plays an important part in determining the overall performance and sensitivity of an antibody, their influence on experimental results will largely depend upon the context in which the antibody is used.
Some researchers might consider sensitivity, which is related to the ability to detect lower abundance antigens, to be a property of an antibody. For example, when characterizing endogenous expression levels of the target in a tissue model, it makes sense to select the more sensitive of two antibodies, while in a cell line model where the target is overexpressed, sensitivity is less of a concern. However, it's more accurate to say that sensitivity is a property of the immunoassay rather than the antibody. This is where factors such as the presentation and concentration of the antigen, buffer conditions (native versus denaturing), temperature, time, and antibody amount come in. An antibody that is highly specific in one immunoassay may exhibit higher cross-reactivity and lower specificity in a different immunoassay. Likewise, an antibody from one vendor might exhibit higher sensitivity in ELISA than an antibody from a second vendor, but the second antibody exhibits higher sensitivity in immunohistochemistry.
All these variables can impact affinity, avidity, specificity, and selectivity, and consequently antibody sensitivity and performance in a given immunoassay. This makes it critical that you confirm that each antibody you use in your experiment has been tested, validated, and optimized for your model system and application before embarking on hypothesis-driven research.
Read the entire Antibody Essentials series:
- Antibody Essentials Part 1: Antibody Basics
- Antibody Essentials Part 2: Antibody Diversity and Classification
- Antibody Essentials Part 3: How Antibody Technologies Evolved
- Antibody Essentials Part 4: Polyclonal vs Monoclonal Antibodies
- Antibody Essentials Part 5: Important Considerations for Antibody Selection
Select References
-
Price R. (2017) How the Reproducibility Crisis in Academia is Affecting Scientific Research. Forbes.com
-
Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521(7552):274-276. doi:10.1038/521274a
-
Baker M. 1,500 scientists lift the lid on reproducibility. Nature. 2016;533(7604):452-454. doi:10.1038/533452a
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531-533. Published 2012 Mar 28. doi:10.1038/483531a







