3D cell culture models are transforming biological research. Unlike 2D assays, where cells are grown on flat, rigid surfaces, 3D cultures allow cells to interact in all directions and form structures that more closely resemble real tissues. These systems better capture the complexity of the cellular microenvironment, ultimately providing more relevant data for translating laboratory findings to clinical settings.
 |
 |
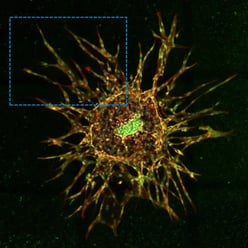 |
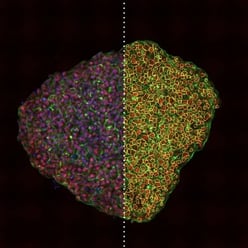
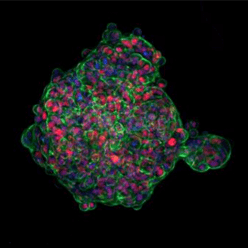
| Figure 1. Three different 3D cell cultures analyzed using CST® antibodies. Left: Scaffold-free, suspension-cultured spheroid analyzed using SMAD2/3 (D7G7) Rabbit Monoclonal Antibody #8685 (red). Center: Matrix-embedded spheroid model seeded in collagen and analyzed using Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit Monoclonal Antibody #18338 (red). Right: 3D sprouting angiogenesis spheroid grown in a fibrinogen matrix and analyzed using SMAD4 (D3R4N) Rabbit Monoclonal Antibody #46535 (red). |
||
Adding urgency to this shift, in 2025, the US Food and Drug Administration (FDA) announced a plan to phase out animal testing requirements for monoclonal antibodies and other drugs. US regulators now formally recognize New Approach Methodologies (NAMs) such as 3D cell cultures, organoids, and organ-on-a-chip technologies as scientifically valid, human-relevant models for drug discovery.
“Culturing cells in 3D rather than in 2D enables experiments to be performed in more physiologically relevant model systems," explains Frank He, PhD, Applications Product Manager at CST. "In traditional 2D culture, cells grow as a monolayer on a stiff plastic surface. In 3D, they interact more extensively with each other and the extracellular matrix. This could have a significant impact on cell signaling pathways and drug response."
Even as 3D models become more accessible and cost-effective, their development and optimization still require significant upfront investment time. While many labs already have the required instrumentation, researchers must overcome a number of practical and technical challenges.
Implementing 3D Cell Culture Models for Drug Discovery
While less complex than organoid or organ-on-a-chip models, 3D spheroid and matrix-embedded cell culture models present their own unique challenges. They are less standardized than 2D models, making protocol optimization and quantification more demanding. Researchers often need advanced imaging and computational analysis to accurately measure protein expression or signaling events.
Greg Innocenti, Associate Director of Immunofluorescence at CST, notes: “Although a growing number of 3D culture formats are now readily available, actually integrating them into laboratory workflows is challenging. Researchers must determine which 3D cell culture microenvironment is most relevant, decide which metrics to track, and adapt the cultures for existing high-content screening workflows.”
Beyond integration issues, reproducibility remains a significant hurdle. Innocenti adds:
“The lack of standardized protocols can make it difficult to reproduce results between experiments and across labs. Issues like light scattering, autofluorescence from scaffolds (e.g., collagen), and the difficulty of achieving uniform staining throughout a 3D structure can compromise data quality. And analyzing the large, multi-layered datasets generated from 3D imaging is a significant bottleneck for many labs."
Additionally, antibodies developed for use in flat, 2D monolayers may not necessarily translate when used in more complex, matrix-embedded structures. Reducing risk wherever possible is a key part of successful assay design and optimization.
"There can be a lot of challenges involved in getting started with 3D models. Having to wrestle with improperly validated reagents isn’t a variable any researcher wants to contend with,” explains Innocenti.
Rigorously validated antibodies help reduce experimental risk and keep project timelines on track. Successful interpretation of immunofluorescence (IF) results in 3D cultures requires the same robust performance and high specificity as is required in 2D systems. CST antibodies are stringently validated in multiple model systems, providing researchers with reliable reagents that are better equipped to detect targets in dense, multi-layered structures with high fidelity.
This proven reliability is why CST remains the most cited antibody provider, trusted by researchers for their most innovative and demanding applications, including 3D cell models.
Antibody Selection for 3D Assays
As 3D cell culture gains popularity in drug screening and high-throughput workflows, choosing the right antibody is critical. Spheroids can be created through scaffold-free methods—such as hanging drop, forced floating, or agitation methods—or scaffold-based techniques using matrices like collagen or gel, including embedded, dome, or sandwich methods. Each format introduces unique considerations for antibody performance and assay design.
One central tenet is true, however: High-quality, rigorously validated antibodies help to ensure signal specificity, sensitivity, and reproducibility, regardless of the assay type.
Antibody Penetration in a Dense, 3D Environment
Antibodies must consistently and uniformly infiltrate the dense, multilayered structures that are characteristic of 3D models. Spheroid or matrix-embedded cultures present unique diffusion barriers that can limit antibody access to target protein epitopes and reduce signal strength. Even an antibody that works well in 2D assays may produce weak or uneven staining in 3D unless it is optimized for effective diffusion and specific target recognition.
For example, in a recent study of epithelial-to-mesenchymal transition (EMT) in cancer malignancy, CST antibodies were used in a multicellular 3D spheroid outgrowth assay (Figure 2) using A549 spheroids seeded in collagen and exposed to TGF-β1. SMAD4 (D3R4N) Rabbit Monoclonal Antibody #46535 was selected because of its rigorous validation across multiple model systems and applications (i.e., #46535 is validated for use in: western blot, immunoprecipitation, immunofluorescence, immunohistochemistry, flow cytometry, and chromatin immunoprecipitation).
 Figure 2. 3D A549 spheroids were cultured on top of a collagen gel for 4 days in the presence/absence of TGF-β1, showing nuclear translocation of total SMAD4. Red: SMAD4 (D3R4N) Rabbit Monoclonal Antibody #46535; Green: F-actin. Bottom panels show an enlarged view of boxed regions in the top panels.
Figure 2. 3D A549 spheroids were cultured on top of a collagen gel for 4 days in the presence/absence of TGF-β1, showing nuclear translocation of total SMAD4. Red: SMAD4 (D3R4N) Rabbit Monoclonal Antibody #46535; Green: F-actin. Bottom panels show an enlarged view of boxed regions in the top panels.
The SMAD4 signal produced by the antibody was robust and uniform, both within the spheroid core and on the periphery. This consistency in staining allowed researchers to clearly and confidently quantify pathway activation and spatial cellular responses in a challenging 3D environment.
To learn more about the 3D EMT model in Figure 2, download the AACR 2024 poster from Agilent and CST.
Highly Sensitive & Specific Antibodies for Robust Dose-Response Analysis
Generating robust dose-response curves in 3D cell culture requires antibodies that can deliver clear, consistent, and quantitative signals throughout all regions of the 3D model. Poor penetration or insufficient specificity translates into weak, uneven staining or variable data, undermining dose-response accuracy.
For example, in a similar spheroid model, A549 lung adenocarcinoma spheroids were seeded in a collagen matrix and cultured in the presence of increasing concentrations of TGF-β1. Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit mAb #18338 was selected because of its rigorous validation across multiple model systems (e.g., FFPE tissue) and applications (i.e., #18338 is validated for use in: WB, IP, IF, F, and ChIP).
 Figure 3. Left: Maximum intensity projections of A549 spheroids treated with vehicle control or 100 ng/mL TGF-β1. Red: Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit Monoclonal Antibody #18338; Green: F-actin; Blue: Nuclei. Right, top: Plate layout of an experiment where triplicate spheroids were treated with decreasing amounts of TGF-β1 (left to right). Right, bottom: Quantified nuclear phosphorylated SMAD2 at each concentration assayed, with an EC50 of 0.31 ng/mL.
Figure 3. Left: Maximum intensity projections of A549 spheroids treated with vehicle control or 100 ng/mL TGF-β1. Red: Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit Monoclonal Antibody #18338; Green: F-actin; Blue: Nuclei. Right, top: Plate layout of an experiment where triplicate spheroids were treated with decreasing amounts of TGF-β1 (left to right). Right, bottom: Quantified nuclear phosphorylated SMAD2 at each concentration assayed, with an EC50 of 0.31 ng/mL.
When analyzed using a high-throughput imaging protocol on the Agilent BioTek Cytation C10 confocal imaging reader, the quantification of TGF-β1-induced phosphorylation and nuclear translocation of SMAD2 in 3D spheroids was consistent and robust.
This reliability empowers researchers to confidently interpret signal changes, relate them to drug effects, and compare findings across both simple and complex systems.
To learn more about the TGF-β1 dose response curve in Figure 3, download the AACR 2024 poster from Agilent and CST.
The CST Antibody Advantage for 3D Immunoassays
For over 20 years, CST antibodies have set the industry standard for reliability and high performance. All regents are extensively validated and tested in relevant model systems—the same model systems used in research labs worldwide. For immunofluorescence reagents, this includes high-content imaging workflows and complex tissue types, giving researchers confidence that their results detect the intended target with high specificity and affinity.
As researchers continue to move toward more physiologically relevant systems, CST antibodies provide the reliability and specificity needed to turn complex imaging data into meaningful biological insights.
Watch the webinar, A Multiscale Approach to Quantitatively Evaluate the SMAD Signaling Pathway, to learn more about assay development and antibody selection for high-throughput and 3D model systems.
Additional Resources
- Blog: High-Throughput 3D Assay Methods: Analysis of TGF-β Signaling
-
Resource Center: High-Content Imaging
-
Application Note: High-Throughput Methods to Quantitatively Evaluate TGF-β Signaling in Angiogenesis
-
Application Note: High-Throughput Methods to Quantitatively Evaluate TGF-β Signaling in Epithelial-to-Mesenchymal Transition




/42157_chimeric%20antibody%20blog%20featured3.webp)