Spheroids and related three-dimensional (3D) cell models are rapidly gaining popularity among researchers due to their ability to more accurately represent in vivo biological features, including tissue architecture, gene expression, and metabolic profiles, thereby surpassing many limitations of traditional 2D cell culture models. This is especially true in cancer research, where cancer spheroid models have been developed to mimic 3D tissue organization within the tumor microenvironment (TME). The densely packed, tumor-like structure of a spheroid and its ability to more accurately represent the complex network of cell-cell contacts within the TME enables contextually relevant modeling of biological processes whose pathologic states represent key hallmarks of cancer.
When used in combination with traditional biochemical and 2D cellular assays, spheroids like the example shown in Figure 1 are another device in the researcher’s toolkit to more comprehensively evaluate biological processes, and they are increasingly being considered as models to assess the efficacy and safety of therapeutics.
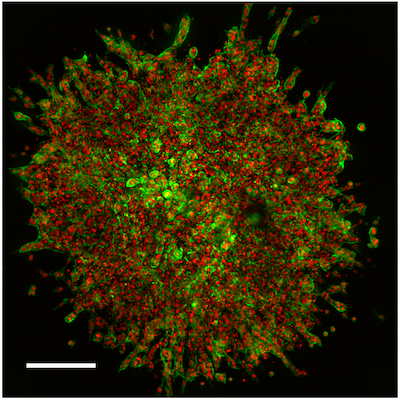 Figure 1. 3D angiogenesis sprouting assay using HUVEC spheroids imaged on the Agilent BioTek Cytation C10 confocal imaging reader. Scale bar corresponds to 200 μm.
Figure 1. 3D angiogenesis sprouting assay using HUVEC spheroids imaged on the Agilent BioTek Cytation C10 confocal imaging reader. Scale bar corresponds to 200 μm.
As high-throughput drug screening campaigns increasingly utilize 3D cellular models, it is essential to ensure that reagents designed to interrogate biological functions within the spheroid are optimized for use in a 3D model context. In the case of immunoassays, both protocols and reagents need to be rigorously tested to ensure effective antibody penetration of the spheroid and specific binding to the target epitope. Factors such as sample preparation, matrix and culture medium compositions, and spheroid size and density are among the many factors that can potentially affect antibody binding performance—and hence the experimental results.
Recently, CST scientists teamed up with Agilent Technologies to demonstrate how CST® antibodies can be used in a variety of assay types on Agilent’s state-of-the-art imager, the Agilent BioTek Cytation C10. To demonstrate the comprehensive and powerful nature of these two technologies working together, the research team evaluated responses to TGF-β signaling pathway activation at different biological scales: Biochemical, 2D cellular, and 3D spheroid models.
The project is summarized below, and you can find additional details in the application note, High-Throughput Methods to Quantitatively Evaluate TGF-β Signaling in Angiogenesis.
Biochemical, 2D and 3D Assay Methods to Evaluate TGF-β Pathway Activation
The transforming growth factor-β (TGF-β) signaling pathway performs essential roles in regulating fundamental cellular processes including proliferation, differentiation, and migration, and as such, plays a critical part in animal development, health, and disease. Pathological alterations to TGF-β signaling have been associated with numerous diseases, including cancer, fibrotic diseases, and pathological immune dysfunction.
Due to its relevance in various disease states, as of 2023, there are approximately 60 active clinical trials involving TGF-β signaling, with nearly 200 additional clinical trials actively recruiting patients.
Evaluating TGF-β Pathway Activation: SMAD Phosphorylation & Translocation
The canonical TGF-β signaling pathway consists of over 30 ligands, which activate receptors belonging to one of two primary signaling axes. One pathway axis is activated through TGF-β ligand-receptor interactions, while the other is activated through BMP ligand-receptor interactions.
Activation of receptors in either pathway axis results in receptor dimerization and autophosphorylation, which leads to recruitment and phosphorylation of pathway-specific receptor-mediated SMAD proteins (R-SMADs), which are the primary signal transduction molecules for the two pathway axes. SMAD protein phosphorylation is therefore used as a robust and reliable readout of pathway activation following ligand-receptor engagement in either axis.
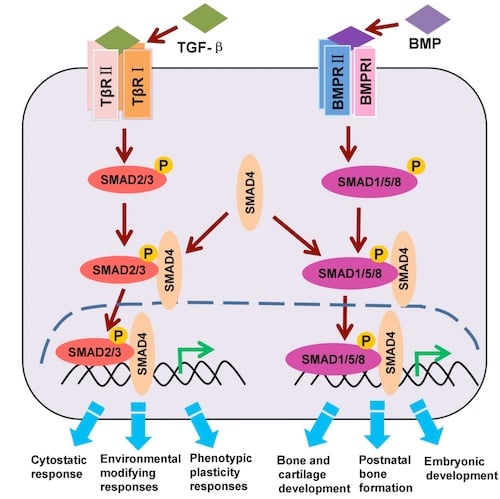 Figure 2. The canonical TGF-β/BMP/SMAD signaling pathway (adaption).
Figure 2. The canonical TGF-β/BMP/SMAD signaling pathway (adaption).
As shown in Figure 2, phosphorylation of SMAD2 and/or SMAD3 indicates activation of the TGF-β pathway axis, while phosphorylation of SMAD1, SMAD5, and/or SMAD9 (sometimes referred to as SMAD8) indicates BMP pathway axis activation. Once phosphorylated, SMAD proteins in either axis recruit a co-SMAD, SMAD4, which then translocates to the nucleus and regulates the transcription of target genes.
Three Assays to Evaluate TGF-β Pathway Activation
Three assay types—biochemical, 2D cellular, and 3D spheroid models—can be used to comprehensively evaluate activation of the TGF-β and BMP signaling pathways. These assays measure the magnitude of SMAD phosphorylation and functional output, both of which are dependent on ligand concentration and exposure duration, and can be used to assess stimulated angiogenesis or reduced cell proliferation.
The Agilent BioTek Cytation C10 is an affordable, benchtop confocal imaging device that combines automated confocal and widefield microscopy with conventional multimode microplate reading. The unique design, which includes a combination of spinning disk confocal and widefield imaging, enables high-resolution imaging and optical sectioning capabilities for many sample types.
 Figure 3. Agilent BioTek Cytation C10 Confocal Imaging Reader.
Figure 3. Agilent BioTek Cytation C10 Confocal Imaging Reader.
Application-specific antibodies from CST and the BioTek Cytation C10 were used to assess TGF-β/BMP signaling, generating the following results:
Biochemical Model
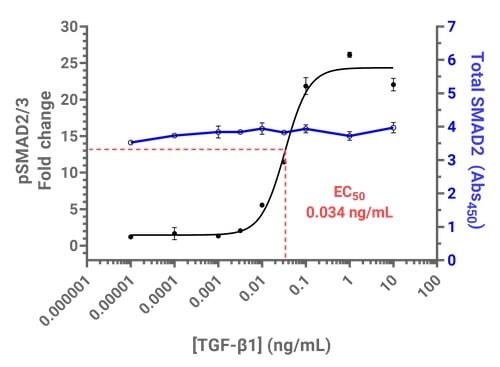 Figure 4. TGF-β1-induced SMAD2/3 phosphorylation can be measured using PathScan® ELISA kits on the Cytation C10. Above: Dose-dependent SMAD2/3 phosphorylation in response to TGF-β using PathScan® Phospho-SMAD2 (Ser465/467)/SMAD3 (Ser423/425) Sandwich ELISA Kit and PathScan® Total SMAD2/3 Sandwich ELISA Kit #12000.
Figure 4. TGF-β1-induced SMAD2/3 phosphorylation can be measured using PathScan® ELISA kits on the Cytation C10. Above: Dose-dependent SMAD2/3 phosphorylation in response to TGF-β using PathScan® Phospho-SMAD2 (Ser465/467)/SMAD3 (Ser423/425) Sandwich ELISA Kit and PathScan® Total SMAD2/3 Sandwich ELISA Kit #12000.
2D Cellular Model

Figure 5. Phospho-specific SMAD antibodies and widefield imaging enable high-throughput quantification of TGF-β1-induced SMAD2/3 phosphorylation and nuclear translocation in 2D cell cultures. Above: Cells treated with vehicle control (A) or TGF-β1 (B) fixed and immunostained using Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit mAb #18338 (green). Scale bars correspond to 100 μm.
3D Spheroid Model
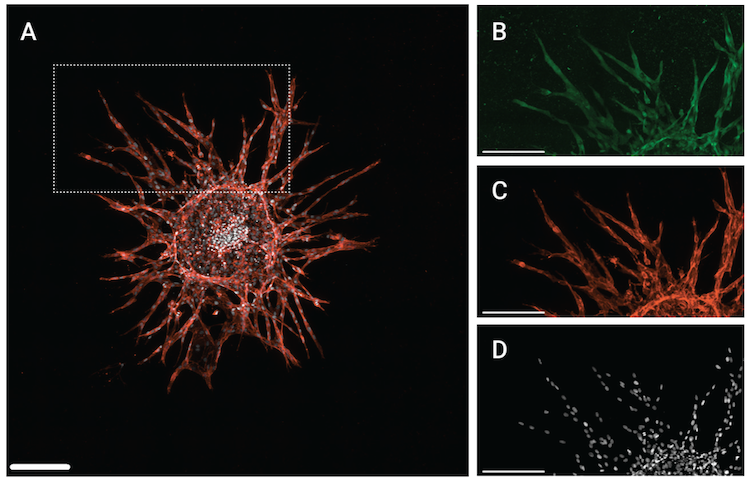
Figure 6. SMAD4 expression was measured using a 3D sprouting angiogenesis model. Above: (A) Maximum projection from a 3 x 3 montage Z-stack of a HUVEC spheroid with angiogenic sprouts. (B) to (D) show the inset region in (A) with the signal for (B) SMAD4 using SMAD4 (D3R4N) XP® Rabbit mAb #46535 (C) PECAM-1 using CD31 (PECAM-1) (D8V9E) XP® Rabbit mAb #77699, and (D) DRAQ7 using DRAQ7 #7406. Scale bars correspond to 200 μm.
As shown in the figures above, a SMAD4 antibody was used to evaluate pathway activation in the spheroid model, while phospho-SMAD antibodies were used in the biochemical and 2D cellular models. This is because the SMAD4 antibody generated the best signal-to-noise ratio in the 3D model compared to the phospho-SMAD antibodies, demonstrating that not all reagents perform equally in all contexts. Since SMAD4 is recruited as a co-SMAD in both the TGF-β and BMP pathways, it serves as a useful marker of SMAD complex localization/translocation.
Antibodies for Immunostaining in 3D Models
In vitro model systems are foundational tools required to understand biological processes, but they have limitations in the extent to which they can replicate in vivo biology. Incorporating 3D spheroid models that better reflect in vivo tissue architecture can provide more accurate screening environments for identifying potential therapeutic targets and assessing drug safety.
Leveraging high-quality, application-specific antibodies, along with state-of-the-art imaging technology, can provide the tools necessary to quantitatively assess biological processes and drive the development of novel therapeutics.
Additional Resources
- Poster: A Multiscale Approach to Quantitatively Evaluate the SMAD Signaling Pathway
- Webinar: Quantitative Evaluation of Biomarkers for TGF-β-Induced EMT in Biochemical, Cellular, and 3D Spheroid Model Systems
- SLAS2023 Video Presentation: A Multiscale Approach to Quantitatively Evaluate the SMAD Signaling Pathway
- Speakers: Antony Wood, PhD, Senior Director, Product Design & Strategy at CST, and Ernest Heimsath, PhD, Applications Development Scientist at Agilent Technologies
- Interactive Pathway Diagrams:
Antony (Tony) Wood, PhD, Senior Director, Product Design & Strategy at CST, contributed to the writing of this post.23-BPA-25500








