Although coronaviruses have long circulated throughout human populations, the study of these viruses has intensified over the last two decades, due to the rise of novel coronaviruses that have greatly impacted human health. The emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, Middle East respiratory syndrome-related coronavirus (MERS-CoV) in 2012, and the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, has emphasized the importance of understanding the fundamental mechanisms by which these viruses cause disease.
SARS and SARS-CoV-2 initiate cell entry by binding to the angiotensin-converting enzyme 2 (ACE2) protein via their trimeric spike (S) glycoprotein. This spike protein comprises two functional subunits, S1 and S2. The S1 subunit encompasses four domains specified as S1A-S1D and is involved in binding to the host cell receptor attachment to the cell; S2 is utilized for fusion between the viral and cellular membranes. The S1B domain, or Receptor Binding Domain (RBD), is of particular importance, as this is the domain that directly binds to ACE2 and facilitates membrane fusion (1).
Recombinant Proteins for Studying SARS-CoV-2
Cell Signaling Technology (CST) has developed a collection of recombinant proteins, corresponding to different domains of the SARS-CoV-2 Spike protein and the host receptor ACE2, that can be used to study COVID-19. The proteins are available with different tag configurations (e.g., 8xHis, murine Fc), allowing for the flexibility to be used in a variety of assay formats.
Researchers can therefore take advantage of the multiple Spike S1-NTD recombinant proteins, which are critical for studying SARS-CoV-2 interactions with ACE2.
 The purity of SARS-CoV-2 Spike S1-NTD (16-316) Recombinant Protein (mFc-Tag) #61516 was determined by densitometry after SDS-PAGE of 2 μg of protein followed by staining with Coomassie Blue. Purity values were determined from DTT-reduced samples (+).
The purity of SARS-CoV-2 Spike S1-NTD (16-316) Recombinant Protein (mFc-Tag) #61516 was determined by densitometry after SDS-PAGE of 2 μg of protein followed by staining with Coomassie Blue. Purity values were determined from DTT-reduced samples (+).
CST offers a recombinant protein corresponding to the full-length Spike protein ectodomain engineered to express as a trimer, which reflects the native configuration of the Spike protein on the viral surface, as well as multiple forms of the Spike receptor-binding domain (RBD) protein, enabling substantial flexibility in assay design.
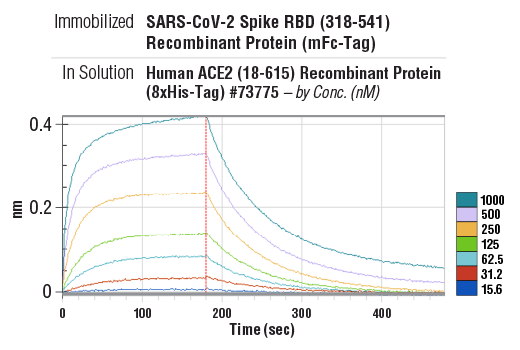
Binding kinetics between SARS-CoV-2 Spike RBD (318-541) Recombinant Protein (mFc-Tag) (immobilized) #41701 and Human ACE2 (18-615) Recombinant Protein (8xHis-Tag) #73775 (in solution, at indicated concentrations). The vertical red line (180 sec) indicates addition of PBS to induce dissociation. Binding was detected with an anti-mouse Fc biosensor. Values on y-axis indicate binding response signals recorded for 7 different concentrations of Human ACE2 (18-615) Recombinant Protein (8xHis-Tag) #73775 (15.6, 31.2, 62.5, 125, 250, 500 and 1000 nM).
The trimeric protein can be used as a component of an ELISA assay for detecting COVID-19 seroconversion (2), which allows for a quick and reliable measurement of SARS-CoV-2 antibody titers. The SARS-CoV-2 Spike RBD (multimeric) (319-591) Recombinant Protein (8xHis-Tag) #17862, a specially engineered version of the spike protein RBD, has demonstrated a higher avidity for the ACE2 receptor than RBD proteins expressed recombinantly as a monomer.
CST has also generated a biotinylated form of the multimeric spike RBD, which offers a reproducible and predictable way to attach the protein to a surface across many different assay platforms. This makes it an ideal option for inhibitory assays where the goal is to prevent the binding between the SARS-CoV-2 spike protein and ACE2 (3) thereby preventing viral entry into the cell. The ACE2 and spike RBD recombinant proteins may also be used in the development of peptide inhibitors, characterizing antibody cross-reactivity, or examining binding kinetics between the virus and the host receptors.
Tools to Help Researchers Fight COVID-19
CST has well-validated research antibodies and small-molecule compounds, which can help researchers unravel the pathogenesis of viral disease and advance the research and development of assays and therapeutic drugs.
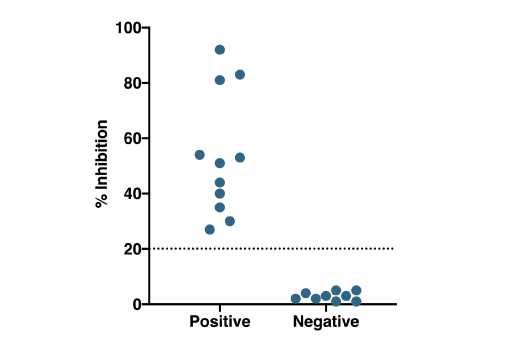
Patient Testing: Patient samples were tested using the SARS-CoV-2 Spike RBD-ACE2 Blocking Antibody Detection ELISA Kit #69486. Serum/plasma samples were heat-inactivated (56°C for 30 min) and diluted 1:10 prior to running the assay, as described in the protocol. Percent inhibition of the SARS-CoV-2 spike RBD-ACE2 interaction (% Inhibition) is plotted for each serum/plasma sample, with values corresponding to samples obtained from donors who had been diagnosed with SARS-CoV-2 (diagnosed positive n=11*) on the left, and values corresponding to samples from presumed uninfected donors.
Antibodies Against the Host Protein
| Target | Product | Note |
| ACE2 | ACE2 Antibody #4355 | Host receptor of SARS-CoV-2 and SARS-CoV |
| DPP4 | Host receptor of MERS-CoV | |
| AAK1 | AAK1 Antibody #79832 | Host endocytosis regulator |
| Cathepsin B | Possibly participates in the hydrolysis of coronavirus S protein, releasing its nucleic acid |
|
| IgM | IgM antibody #74293 |
Can be used with SARS-CoV-2 specific antigen to detect whether there is SARS-CoV-2 specific IgM in patients' blood. |
Candidate Compounds to Help Understand Drug Effects
Product |
Note |
| Chloroquine #14774 | Lysosophilic agent which inhibits autophagy and can inhibit SARS-CoV-2 in vitro[6] |
| Sunitinib #12328 | Multiple tyrosine kinase inhibitors targeting PDGFR, VEGFR, KIT and FLT3 |
| Erlotinib #5083 | ATP Competitive Inhibitor in EGFR Kinase Pathway |
| Carfilzomib #1502 | Epoxomycin-related protease inhibitor |
Given the potential for inducing multiple severe conditions, scrutinizing SARS-CoV-2, its protein components, and its interactions with ACE2 is of critical importance. Recombinant proteins toward these components are extremely useful reagents to those undertaking this research, which will ultimately provide us with the tools needed to identify and combat these viral infections.
References:
- Wang, C. et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Commun. 11, 1–6 (2020).
- Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Med. (2020). doi:10.1038/s41591-020-0913-5
- Nie, J. et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Microbes Infect. 9, 680–686 (2020).