Whether your heart is devoted to cardiac research, you’re excited to investigate neurodegenerative diseases, you’re immune to the trials and tribulations of infectious disease research, or you’re willing to stop at nothing to halt cancer cells in their tracks, we’ve all needed to answer this deceptively easy question: how much of (insert name of favorite target) protein is in my samples?
Each target and experimental question needs a measurement method that best fits that specific circumstance, of which several factors need to be considered: What is the overall scientific question? How many and what type of samples am I running? Am I looking at one target or many? Do I need a rough/qualitative measurement or a very accurate/quantitative measurement? Do I need a highly sensitive assay? What is the dynamic range of the target across samples? Does this project warrant months of upfront work of setup, or would I be better off using a simpler/easier method? Do I even have the equipment to run this!?
These methods range widely, each with their own set of strengths and weaknesses.
Mass Spectrometry vs Antibody-Based Detection
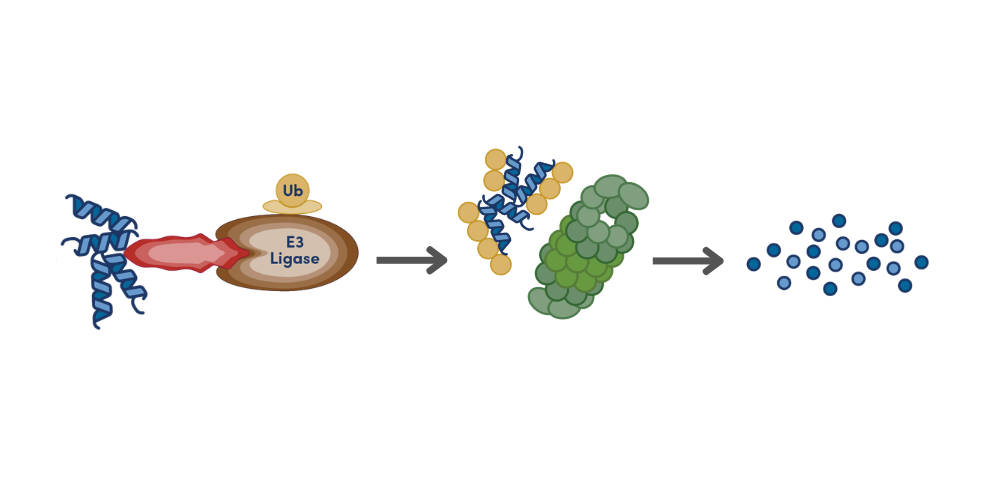
There are methods that directly measure your protein of interest like mass spectrometry. This can be used for discovery projects when global proteomic measurements are desired, or for sensitive quantitation of select proteins when using targeted proteomic techniques. Although an extremely powerful tool, mass spectrometry generally requires complex equipment, workflows, sample preparation, and analysis compared to other methods.
Most other analysis methods indirectly measure your target via a binding event(s) using a protein binder (typically an antibody) that is specific for your target protein.
Examples of this include the traditional western blot, which allows for basic visualization of protein levels, and is a technique that most lab researchers are familiar with. If additional information (such as localization or cell type expression) is needed, more complex techniques like flow cytometry and IHC/IF can be used. These are especially useful when your sample type is a heterogeneous population of immune cells or is composed of various cell types (such as in tissue), where changes of your protein may only occur in a specific cell type. These techniques shine in many circumstances but come with tradeoffs, such as lower throughput (Western Blot), and are not as quantitative as other techniques when measuring overall target levels (flow cytometry, IF, IHC).
Advantages and Limitations of ELISA for Protein Quantitation
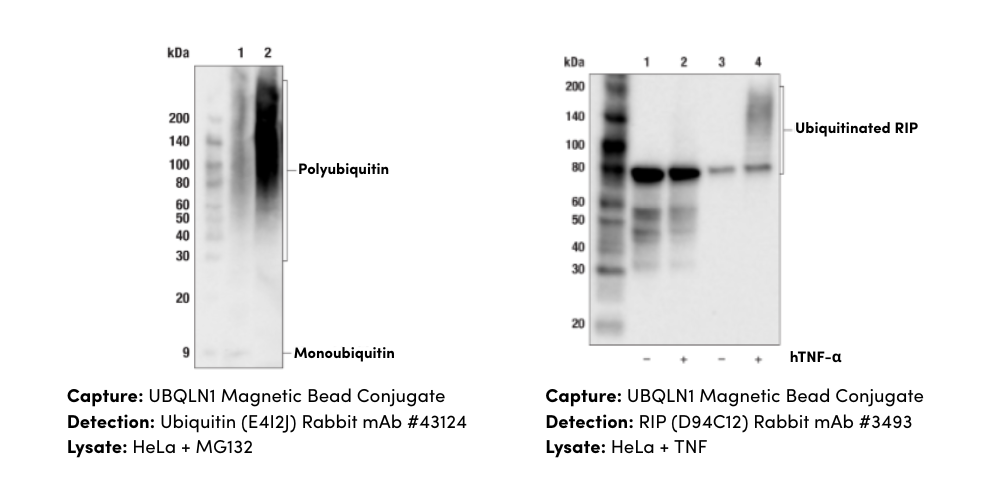
When your main objective is strictly sensitive quantitation of a protein in large numbers of samples (cell/tissue extract, serum, or other biofluid), immunoassays such as ELISA are a great fit. While there are many flavors of immunoassays, sandwich ELISAs are the workhorses commonly used in the lab to measure protein levels by forming a complex of a protein “sandwiched” between two antibodies specific to different epitopes of that target and detected using an enzyme-conjugate.
ELISAs are a frequently used platform because they are simple and quick to run. Where ELISAs really shine are in screening and dose-response experiments (compound screens, inhibition curves for therapeutic compounds, biomarker measurements in patient samples during disease progression or after therapeutic intervention) where large numbers of samples are being run and straightforward and accurate quantitation (relative or absolute if paired with a protein standard) is necessary.
ELISAs can easily handle large sample numbers since they are usually set up as 96 or 384-well assays, but can be higher-throughput if paired with robotic automation. Most importantly, sandwich ELISAs are considered the gold standard for quantitation—they are typically more sensitive with a larger dynamic range than other techniques (such as western blot) while still maintaining high specificity. These benefits do come with a tradeoff—sandwich ELISAs require the availability of high-quality antibodies that work as a pair, and they require more time and effort upfront to make, optimize, and validate. In addition, traditional sandwich ELISAs are not ideally suited for highly multiplexed setups. Although they’re great for protein quantitation in bulk samples (biofluid, cell/tissue extract), if localization or quantitation in specific cell types is also required, you’ll need to incorporate additional methods into your workflow.
Emerging Technologies in ELISA-Like Assays
Newer ELISA-like assay technologies can leverage antibodies/antibody pairs used in traditional ELISA to further build on the quantitative strengths of traditional ELISA while also adding capabilities, although this usually requires specialized equipment and a more complex setup. For example, advances in detection methods such as label-free detection (surface plasmon resonance, graphene sensors that don’t require antibodies to be labeled with an enzyme or fluorophore) and label-based detection (fluorophores, DNA-based detection, proximity-induced detection), as well as alternative capture antibody immobilization strategies (bead-based, spot-specific) can allow for additional increases in sensitivity, dynamic range, throughput, assay speed, and multiplexing.
The technology for ELISA-like assays is constantly evolving, with newer technologies even going in the direction of single-cell and multi-omic measurements (protein, RNA, and DNA).
High-Quality Antibodies Matter for Any Assay
Regardless of how simple or complex your ELISA format is, the bottom line is that all these still require highly specific, sensitive, and validated antibodies/antibody pairs as a foundation for the assay.
Learn more about CST solutions for antibody pair-based immunoassay development: