Twenty years ago, when considering the tools needed for more robust proteomics research to probe cellular signaling, CST scientists asked an innovative question: What if antibodies could bind to post-translational modifications (PTMs), regardless of the flanking amino acid sequence?
This question—and the resulting method developed by CST—is the basis of the PTMScan® line of products and CST® proteomics services, which simplify proteomics analysis by allowing researchers to rapidly identify and quantify thousands of novel PTM sites using liquid chromatography with tandem mass spectrometry (LC-MS/MS).
This post explores how these revolutionary antibodies are produced and some of the groundbreaking studies conducted using this technology.
Finding the Achilles Heel of Proteomics Research
At the turn of the millennium, scientists at CST were grappling with a hurdle many researchers faced. The existing methods for PTM analysis suffered from a lack of sensitivity, specificity, and throughput, and tended to focus on the more abundant proteins present in a sample. This could mean overlooking signaling pathways with proteins that have low expression levels, but that are still critical to disease progression.
For drug development, mutation analyses, and kinase profiling studies, CST scientists turned the traditional antibody development process on its head and devised a new PTM detection method that is still driving crucial discoveries today: PTMScan.
A Novel Method to Detect PTMs: What Is PTMScan?
PTMScan uses a unique approach to antibody development that enables the detection of thousands of novel sites of PTMs in a single LC-MS/MS run. The technology has been successfully deployed to monitor phosphorylation, ubiquitination, acetylation, and other widely researched PTMs.
When creating PTMScan, CST scientists were seeking a way to find aberrant tyrosine kinase activity in cancer cells.1 Activated tyrosine kinases are known drivers of oncogenic signaling, but it can be challenging to identify specific substrates due to the complexity of the signaling pathways and low levels of tyrosine phosphorylation (<1% of the total phosphoproteome). At the time, antibodies could be developed for known tyrosine phosphorylation events, but what about identifying novel sites?
Developing Antibodies to Detect Novel PTM Sites
First, let’s consider how scientists typically make antibodies. Antibodies are commonly made by challenging a rabbit’s immune system with a peptide antigen in order to identify an antigen-specific antibody. This results in a site-specific PTM antibody that has an affinity to a single peptide sequence or protein containing the PTM at a specific amino acid residue within the target sequence. In the example below, a single phosphopeptide is used as the antigen, and an antibody is developed that is specific only to a peptide sequence with the defined phosphotyrosine residue.
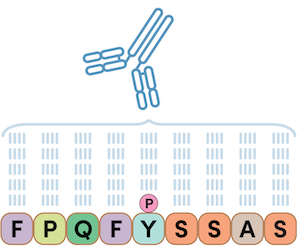
Figure 1. When injected into a rabbit, a phosphopeptide antigen can create an antibody that is specific to a phosphorylated peptide sequence; in the example above, a site-specific phosphotyrosine antibody. The PTMScan Direct line of products is based on this detection technology.
With PTMScan, instead of an antibody specific to a single phosphopeptide sequence, a peptide-library-based approach is used for antibody development, which generates antibodies specific to just the phosphorylated amino acid residue (or any other PTM residue). This strategy can be adapted to create antibodies specific to other post-translationally modified residues (i.e., acetyl-lysine, methyl-arginine, etc.). In this approach, rabbits are immunized using a library of peptides to obtain the phosphotyrosine-specific antibody, where the phosphotyrosine site is fixed and all other flanking positions are randomized amino acids1.
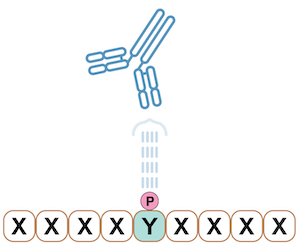 Figure 2. Peptide library example where the phosphotyrosine site is fixed and the flanking positions (marked X) are randomized amino acids. The above example shows a phosphotyrosine-specific antibody. This technology is used to create the PTMScan Discovery product line.
Figure 2. Peptide library example where the phosphotyrosine site is fixed and the flanking positions (marked X) are randomized amino acids. The above example shows a phosphotyrosine-specific antibody. This technology is used to create the PTMScan Discovery product line.
With an antibody specific to any phosphotyrosine site, all peptides with a phosphorylated tyrosine can be immunoprecipitated from a complex mixture after digestion and subsequently identified using LC-MS/MS.
The PTM-specific antibody approach was further extended to consider phosphorylated substrate motifs. In this approach, rabbits are immunized using a library of peptides, where the phosphoserine site and the conserved substrate motif are fixed, while the remaining flanking amino acids are randomized1.
The example below shows a motif antibody where the arginine residue has a fixed position of negative three relative to the phosphoserine. In the peptide library, these two residues are fixed, and the amino acids around those positions vary. This motif antibody will recognize the sequence RXXpS in any protein only when the serine residue is phosphorylated, which is specific to the AKT substrate motif highlighted in the signaling of the AKT pathway.
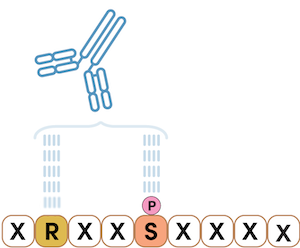
Figure 3. Having just two positions fixed defines the substrate motif RXXpS, where X can be any amino acid. In this example, the arginine residue has a fixed position of negative three relative to the phosphoserine. Motif antibodies are also used for PTMScan Discovery.
Using this method, antibodies can be developed that are exquisitely specific to any targeted modification motif, but broadly reactive against a wide array of different primary amino acid sequences.
CST Products for Bottom-up Proteomics
Using the method for antibody development outlined above, CST offers a suite of PTMScan products that help simplify proteomics analysis for targeted research and initial discovery efforts to efficiently hone in on identifying critical PTMs that drive signaling within your model system.
PTMScan Direct
For targeted studies, PTMScan Direct identifies on- and off-targets in early drug discovery to help de-risk the drug development pipeline. The technology allows for the targeted, pathway-centric screening of known PTMs on protein targets within a selected group of signaling pathways, and can be used to monitor cellular response to a drug treatment or to identify drivers of a specific disease state. PTMScan Direct enables the identification and quantification of hundreds of signaling nodes derived from specific, representative proteins in signaling pathways or cellular compartments.
The PTMScan Multi-Pathway Enrichment kit provides a robust method to identify and quantitatively profile >4000 unique sites and >800 key proteins with known functions in a single experiment.
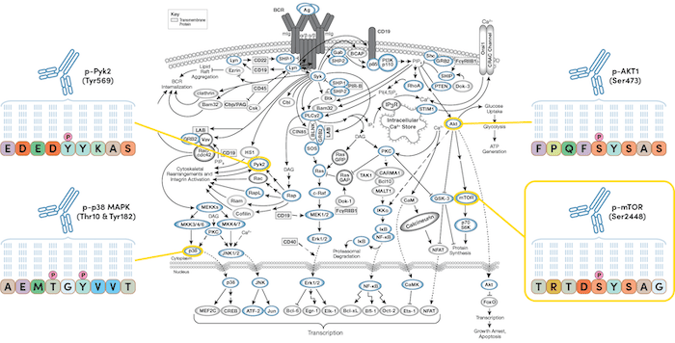
Figure 4. PTMScan Direct can be used to interrogate known PTMs on protein targets within a selected group of signaling pathways.
PTMScan Discovery
PTMScan Discovery, used for discovery-mode studies, searches the entire proteome to identify and quantify your PTM of interest (Figure 5).
Carried out using the PTMScan Motif Antibody kits, PTMScan Discovery identifies critical PTMs throughout the proteome that are modulated between control and perturbed conditions to reveal key sites on proteins that are engaged in response to the test condition(s) or responsible for the progression of the disease (or phenotype).
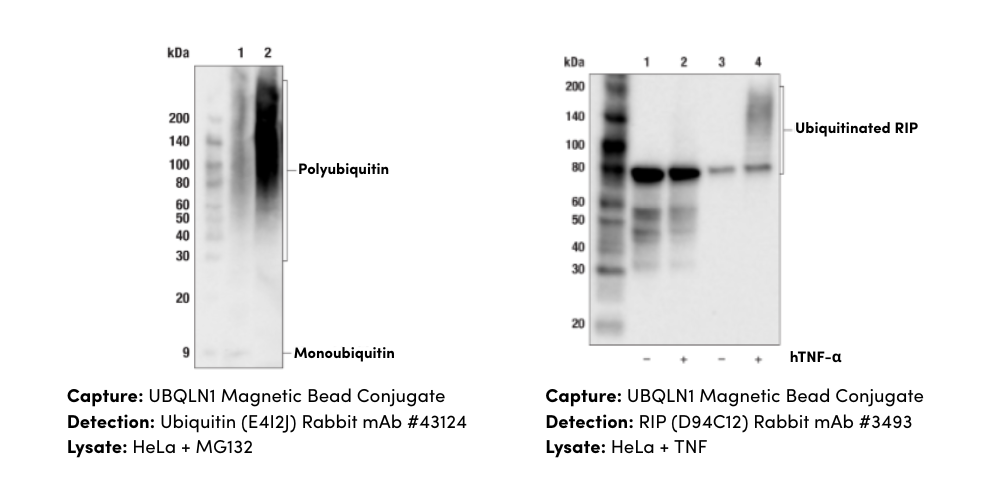
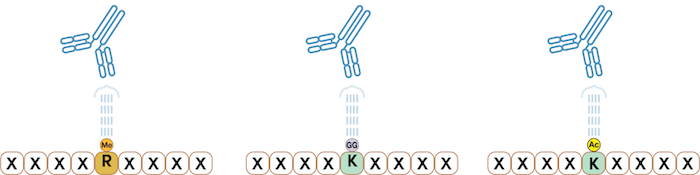 Figure 5. PTMScan Discovery products can detect a specific PTM of interest in your sample. Left: Methyl-Arginine Specific Antibody. Center: Ubiquitin/SUMO-Remnant Specific Antibody. Right: Acetyl-Lysine Specific Antibody.
Figure 5. PTMScan Discovery products can detect a specific PTM of interest in your sample. Left: Methyl-Arginine Specific Antibody. Center: Ubiquitin/SUMO-Remnant Specific Antibody. Right: Acetyl-Lysine Specific Antibody.
PTMScan HS: Proteomics Discovery Using Less Sample
To increase the sensitivity and specificity of the PTMScan reagent, CST scientists leveraged their expertise to produce a PTMScan antibody:magnetic bead conjugate material that results in significantly less antibody protein released from the beads during acid elution. This new PTMScan line of products, PTMScan HS for High Sensitivity, also includes new bind and wash buffers to aid in reducing background peptide binding. Importantly, these kits require less sample, and the magnetic bead format substantially reduces the workload at the bench and makes them amenable to automation.2
CST Proteomics: PTMScan Services
For researchers that do not have access to mass spectrometry equipment but would still like to integrate discovery or targeted PTM studies in their research efforts, the CST Proteomics Services team can serve as an extension of your laboratory to perform qualitative and quantitative profiling of proteins or sites of PTMs in your samples using the technologies outlined above. CST scientists consult with you throughout the project from initial planning to delivery of a comprehensive data package, ensuring your research goals are achieved.
Discoveries Using PTMScan Technology
After its development, the PTMScan technology quickly led to the discovery of novel activators of tyrosine phosphorylation in lung cancer,3 ovarian cancer,4 and acute myeloid leukemia,5 among others. Further commercialization of PTMScan kits empowered scientists from laboratories around the world to identify novel sites of tyrosine phosphorylation and their upstream kinases. Now, oncologists had the information to match patients with known inhibitors to the kinases driving proliferation.
CST scientists soon developed PTMScan antibodies to lysine acylation (acetyl, butyryl, propionyl, and malonyl) and reagents for the identification of ubiquitinated and SUMOylated sites on proteins. PTMScan studies using the unique antibody to the ubiquitin remnant, the motif generated with trypsin digestion, have led to the identification and discovery of tens of thousands of ubiquitination sites on thousands of proteins. Moreover, these studies have revealed the mechanism of action of Lenalidomide in the treatment of multiple myeloma,6 the regulation of pluripotency and cellular reprogramming,7 the requirement for ubiquitination in the invasive phenotype of herpesvirus,8 and substrates of ubiquitin ligase in arthritis.9
More recently, scientists studying the targets of arginine methyl transferase inhibitors in melanoma cells found significant reductions in the methylation of mRNA splicing factors.10 The inhibitors to methyl transferases had previously been shown to slow the growth of some cancer cells. The laboratories employed novel PTMScan reagents designed to enrich for sites of arginine methylation and uncovered another chink in the armor of cancer cells. These splicing factors require methylation by PRMT5 on key arginine residues to properly splice mRNA. Some of the mis-spliced messages encode proteins like MDM4, which are critical in the regulation of p53 and the cell cycle. These, and similar experiments using PTMScan, have identified the proteins key to promoting the growth of cancer cells.
PhosphoSite: An Online Database of PTMs
Concurrent with the invention of PTMScan, CST scientists also created PhosphoSitePlus.org, a curated online searchable database of protein phosphorylation. Phosphosite is populated with published data of protein phosphorylation combined with results from our in-house research, resulting in an online database of PTMs.11,12
PhosphoSitePlus has since expanded to include protein ubiquitination, acetylation, lysine methylation, arginine methylation, and N-glycosylation, allowing researchers from around the world access to information on hundreds of thousands of PTMs.
20 Years of Revolutionary Proteomics Research
Scientists around the world have published well over 100 peer-reviewed manuscripts employing PTMScan. The power of PTMScan derives from leveraging the synergy of three technologies: novel PTM-specific antibodies, cutting-edge mass spectrometry, and bioinformatics. This synergy allows for the identification and relative quantitation of thousands of sites of PTM in one experiment. The promise of personalized medicine depends on identifying the cause of aberrant growth at the molecular level. So twenty years on, with the unbiased approach toward the enrichment of PTM peptides, PTMScan technology still excels as a tool of discovery.
Learn more about proteomics at CST
- PTMScan kits are available for purchase from the CST website, including PTMScan Pathway-Based Enrichment Kits and PTMScan Motif Antibody Kits.
- CST also offers Proteomics Analytical Services using PTMScan.
- Blog: 3x the Data, 3x the Insights: What's the difference between DIA & DDA proteomics?
- Blog: Reproducibility in Proteomics Experiments: Using Control Peptides with PTMScan
Select References
- Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94-101. doi:10.1038/nbt1046
- Rivera KD, Olive ME, Bergstrom EJ, et al. Automating UbiFast for High-throughput and Multiplexed Ubiquitin Enrichment. Mol Cell Proteomics. 2021;20:100154. doi:10.1016/j.mcpro.2021.100154
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190-1203. doi:10.1016/j.cell.2007.11.025
- Ren H, Zhi-Ping T, Zhu X, Crosby K, Haack H, Ren JM, Beausoleil S, Mortiz A, Innocenti G, Rush J, Zhou X, Gu TL, Yang YF, Comb M. Identification of Anaplastic Lymphoma Kinase as a Potential Therapeutic Target in Ovarian Cancer. Cancer Res (2012) 72 (13) 3312-3323
- Gu TL, Goss VL, Reeves C, et al. Phosphotyrosine profiling identifies the KG-1 cell line as a model for the study of FGFR1 fusions in acute myeloid leukemia. Blood. 2006;108(13):4202-4204. doi:10.1182/blood-2006-06-026666
- Krönke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301-305. doi:10.1126/science.1244851
- Buckley SM, Aranda-Orgilles B, Strikoudis A, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11(6):783-798. doi:10.1016/j.stem.2012.09.011
- Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE, Smith GA. Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A. 2015;112(41):12818-12823. doi:10.1073/pnas.1512559112
- Lee KA, Hammerle LP, Andrews PS, et al. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J Biol Chem. 2011;286(48):41530-41538. doi:10.1074/jbc.M111.248856
- Musiani D, Bok J, Massignani E, et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci Signal. 2019;12(575):eaat8388. Published 2019 Apr 2. doi:10.1126/scisignal.aat8388
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512-D520. doi:10.1093/nar/gku1267
- Johnson JL, Yaron TM, Huntsman EM, et al. An atlas of substrate specificities for the human serine/threonine kinome. Nature. 2023;613(7945):759-766. doi:10.1038/s41586-022-05575-3
CST and Cell Signaling Technology are trademarks or registered trademarks of Cell Signaling Technology, Inc. All other trademarks are the property of their respective owners. Visit cellsignal.com/trademarks for more information. 23-BPA-73950
23-BPA-73950