Accounting for about 80% of adult acute leukemias, acute myeloid leukemia (AML) is an aggressive form of blood cancer that is characterized by the uncontrolled proliferation of clonal hematopoietic stem and progenitor cells.1,2 While the disease is initially almost always responsive to chemotherapy and prognosis has vastly improved over the past few decades, the activation of immune-escape mechanisms by leukemic cells and the emergence of chemo-resistant clones cause relapse events to remain rather high.3-5
However, a new class of drugs that target the AML epigenome is offering hope against this aggressive disease. Menin inhibitors are emerging as a promising therapeutic approach, particularly for AML subtypes with specific genetic abnormalities such as Lysine methyltransferase 2A (KMT2A) rearrangements and Nucleophosmin 1 (NPM1) gene mutations.
How do menin-KMT2A inhibitors work? This blog reviews the mechanisms of action and provides an overview of the new therapeutic strategies for treating AML that are currently in clinical trial, as well as describes key targets and the CST® antibodies that can be used by researchers studying the disease.
<<Jump to the end of this post for a list of relevant CST products>>
The AML Epigenome and Mechanisms of Action
AML is a genetically heterogeneous disease with a very low mutational load. Interestingly, however, these relatively few mutations frequently target chromatin and epigenetic modulators. Moreover, a number of oncogenic fusion proteins that drive leukemogenesis in AML function through aberrant expression or redirected specificity of epigenetic regulators. Therefore, targeting the AML epigenome has emerged as an exciting means for discovering more effective therapies.
 Mechanisms of action for NPM1 mutated (left) and KMT2A rearranged AML (right). Adult patients with NPM1c AML and select co-mutations and/or relapsed/refractory disease are associated with poor prognosis. NPM1c accounts for ~30% of AML cases and the five-year overall survival (OS) rate is ~50%. Adult patients with KMT2Ar AML have poor prognosis with high rates of resistance and relapse following current standards of care. KMT2Ar accounts for ~5-10% of AML cases and the five-year OS rate for KMT2Ar AML is <20%.
Mechanisms of action for NPM1 mutated (left) and KMT2A rearranged AML (right). Adult patients with NPM1c AML and select co-mutations and/or relapsed/refractory disease are associated with poor prognosis. NPM1c accounts for ~30% of AML cases and the five-year overall survival (OS) rate is ~50%. Adult patients with KMT2Ar AML have poor prognosis with high rates of resistance and relapse following current standards of care. KMT2Ar accounts for ~5-10% of AML cases and the five-year OS rate for KMT2Ar AML is <20%.
Menin inhibitors that target transcriptional programs driving leukemogenesis in KMT2A-rearranged and NPM1-mutated AMLs are currently under investigation in clinical trials.1,2 The mechanism of action of this class of inhibitors is based on disrupting the critical interaction between the chromatin adaptor protein menin and the KMT2A complex, which ultimately leads to the differentiation and apoptosis of leukemic cells. Excitingly, results from phase I and II clinical trials in which this new class of drugs has been tested alone and in combination with other synergistic agents show promising outcomes in safety and response rates of pre-treated acute leukemia patients.6,7
Menin Dependency and the KMT2A Rearrangement in AML
KMT2A, also known as Mixed Lineage Leukemia 1 (MLL1), was discovered in 1992 from cloning the gene that is disrupted in human 11q23 leukemias and is the mammalian counterpart of drosophila trithorax.8-12 This proto-oncogene encodes a rather large (500 kDa) methyltransferase that functions as a critical epigenetic co-activator and plays key roles in embryonic development and hematopoiesis. Critically, KMT2A is disrupted and undergoes chromosomal translocations in a subset of acute leukemias. In fact, KMT2A rearrangements (KMT2Ar) occur in approximately 10% of acute leukemias and 70-80% of infant leukemias.13,14 About 70% of therapy-related leukemias also harbor KMT2Ar.15,16
These leukemogenic rearrangements fuse the common N-terminus of the KMT2A protein in frame with more than 80 translocation partner genes, many of which are transcriptional cofactors themselves and include AF4, AF9, and MLLT1/ENL. In KMT2Ar leukemias, these fusion proteins interact with chromatin-associating complexes, including the histone methyltransferase DOT1L and the anchor protein menin, to subsequently upregulate genes critical for hematopoietic cell proliferation and differentiation.17
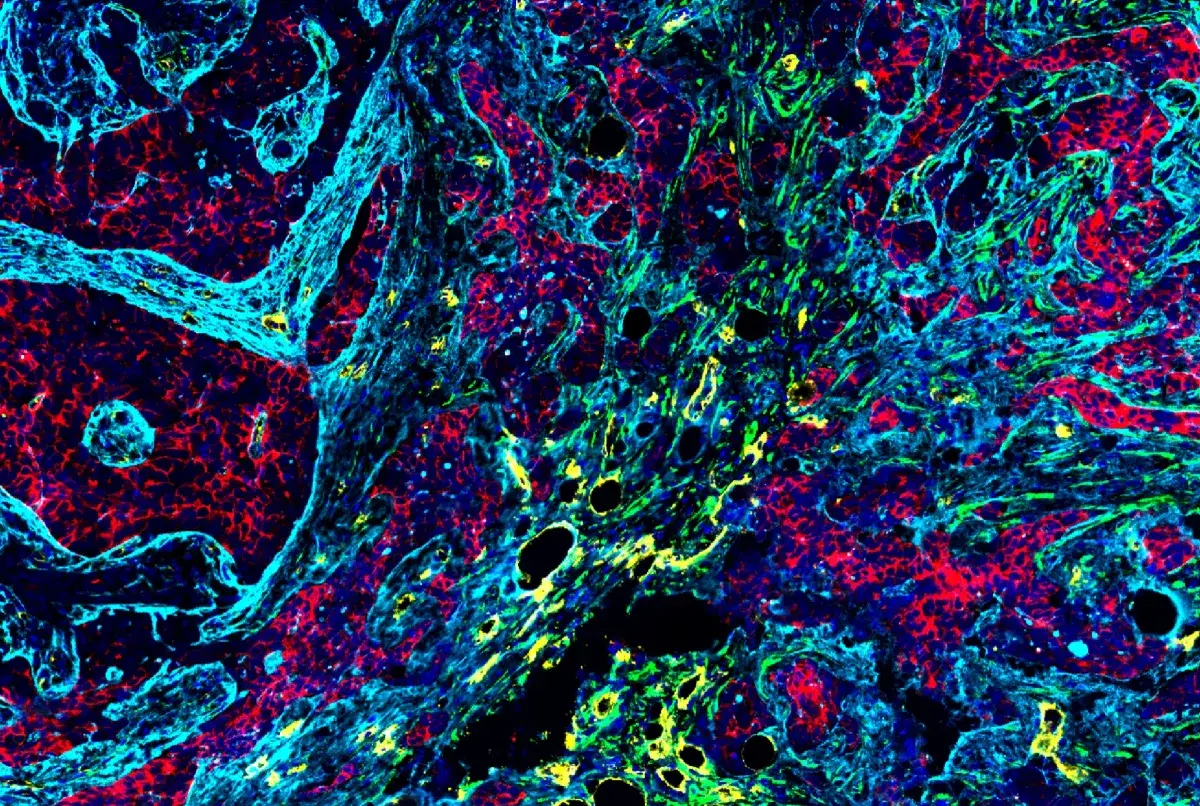
 Confocal immunofluorescent analysis of MEF wild-type (left, positive) and MEF MLL (-/-) (right, negative) cells using MLL (D6G8N) Rabbit mAb (Carboxy-terminal Antigen) #14197 (green). Actin filaments were labeled with DyLight™ 554 Phalloidin #13054 (red). MLL1 WT and KO MEF were kindly provided by Dr. Ali Shilatifard of Northwestern University.
Confocal immunofluorescent analysis of MEF wild-type (left, positive) and MEF MLL (-/-) (right, negative) cells using MLL (D6G8N) Rabbit mAb (Carboxy-terminal Antigen) #14197 (green). Actin filaments were labeled with DyLight™ 554 Phalloidin #13054 (red). MLL1 WT and KO MEF were kindly provided by Dr. Ali Shilatifard of Northwestern University.
Direct therapeutic targeting of the KMT2A fusion proteins is yet to be achieved, but targeting members of the oncogenic fusion complex has shown promising results in attenuating disease progression. Enzymatic inhibition of DOT1L, which is necessary for the development and maintenance of KMT2Ar leukemias, showed great potential for the treatment of these cancers in preclinical studies. However, DOT1L inhibition has only shown modest activity in an early-phase clinical trial when used as a single agent.18 Given menin’s fundamental role in activating leukemic transcriptional programs,19,20 small molecule inhibition of the menin-KMT2A interaction has shown strong therapeutic potential for treating KMT2Ar.21,22 Specifically, menin directly interacts with a highly conserved region within the N-terminal portion of KMT2A that is maintained in all KMT2A-fusion proteins. This interaction facilitates the anchoring of the fusion complex to chromatin, leading to aberrant transcription of key targets such as HOXA9 and MEIS1, which are critical for KMT2Ar AML pathogenesis.23,24
Blocking the menin–KMT2A protein interaction prevents the assembly of KMT2A fusion complexes on chromatin. Preclinical studies have established that menin inhibition downregulates HOXA9 and MEIS1 and reverses leukemogenesis in KMT2Ar leukemia models.21,25 Intriguingly, this class of menin inhibitors also decrease the protein’s total abundance without altering mRNA levels. In KMT2Ar leukemia cells, menin inhibitors promote its interaction with the ubiquitin ligase CHIP, causing increased menin ubiquitination and subsequent degradation.26 While the clinical utility of this degradation is not well understood, as of today, there are at least six different menin-KMT2A inhibitors under clinical evaluation for acute leukemias: DS-1594, BMF-219, JNJ-75276617, DSP-5336, ziftomenib, and revumenib, with promising early patient response data reported for the latter two.6,7
Menin Dependency and the NPM1 Gene Mutation in AML and Other Leukemias
Mutations in the NPM1 gene are also major drivers of leukemogenesis. In fact, they occur in up to 30% of acute leukemias and are the most common genetic alteration in adult AML.27 NPM1 is a chaperone protein that shuttles between the nucleus and the cytoplasm, but primarily is a nuclear protein and carries out various biological functions. The shuttling of NPM1 wild-type is mediated by two nuclear export signals (NES); a bipartite nuclear localization signal and a C-terminal nucleolar localization signal. Importantly, insertions in exon 12 of the NPM1 gene are the most common alterations, and result in the generation of a new NES at the C-terminus, which ultimately leads to stable cytoplasmic delocalization of mutant NPM1 (NPM1c).28,29 Given the pleomorphic roles of NPM1, mislocalization of NPM1c is crucial in promoting leukemogenesis.
The NPM1c AML and KMT2Ar leukemias exhibit similar gene expression profiles, and in NPM1c AMLs, the wild-type KMT2A and menin interaction leads to HOXA9- and MEIS1-driven leukemogenesis.30,31 Similar to KMT2Ar leukemias, menin is also critical for the proliferation and survival of NPM1c AMLs, and utilizing the menin inhibitor has proven successful in destabilizing the wild-type KMT2A complex in NPM1c leukemic models.30,31 Indeed, the ongoing clinical trials of menin inhibitors mentioned above have also been investigating their safety and efficacy in leukemia patients with relapsed or refractory acute leukemias harboring NPM1c with similar promising results.6,7
Future Therapeutic Directions for Mixed Lineage Leukemia Treatment
In summary, menin inhibitors present an exciting new class of drugs against KMT2Ar and NPM1c leukemias and underscore the importance of indirect oncogenic protein complex targeting. In addition, other subsets of leukemias in which HOXA9 and MEIS1 are overexpressed, such as those with nucleoporin 98 (NUP98) rearrangements, might also be responsive to menin inhibition.32 Importantly, identifying rational combination therapies such as BCL-2 and FLT3 inhibitors that appear to demonstrate synergistic effects with menin inhibition will be critical to improve responses and prevent resistance to therapy.
CST Antibodies for Studying AML
For researchers studying menin-MLL inhibitors, CST offers a number of highly validated KMT2A/MLL antibody products. Visit the Cancer Research and Epigenetics Research resource centers on the CST website, and wse the table below to explore relevant products by target and intended application.
| Target | Product | Application(s) |
| Menin | Menin (E5P1R) Rabbit mAb #19893 | ChIP, IHC, WB, IP |
| Menin | Menin (D45B1) XP® Rabbit mAb #6891 | IF, WB |
| DOT1L | DOT1L (D1W4Z) Rabbit mAb #77087 | ChIP, CUT&RUN, WB, IP |
| MLL1/ENL | MLLT1/ENL (D9M4B) Rabbit mAb #14893 | ChIP, CUT&RUN, WB |
| MLL C-Terminal |
MLL1 (D6G8N) Rabbit mAb (Carboxy-terminal Antigen) #14197 | IF, WB, IP |
| MLL N-Terminal |
MLL1 (D2M7U) Rabbit mAb (Amino-terminal Antigen) #14689 | CUT&RUN, CUT&Tag, WB, IP |
| NPM1 | NPM1 (E7W4P) Rabbit mAb #92825 | IHC, IF, WB |
| NPM1c | NPM1 (C Mutant Specific) Antibody #17944 | ChIP, WB, IP |
| NUP98 | NUP98 (C39A3) Rabbit mAb #2598 | IF, WB, IP |
| NUP98 | NUP98 (C37G10) Rabbit mAb #2597 | WB, IP |
| NUP98 | NUP98 (L205) Antibody #2288 | WB |
Select References
- DiNardo CD, Erba HP, Freeman SD, Wei AH. Acute myeloid leukaemia. Lancet. 2023;401(10393):2073-2086. doi:10.1016/S0140-6736(23)00108-3
- Shimony S, Stahl M, Stone RM. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am J Hematol. 2023;98(3):502-526. doi:10.1002/ajh.26822
- Lokody I. Drug resistance: Overcoming resistance in acute myeloid leukaemia treatment. Nat Rev Cancer. 2014;14(7):452-453. doi:10.1038/nrc3776
- Vago L, Perna SK, Zanussi M, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med. 2009;361(5):478-488. doi:10.1056/NEJMoa0811036
- Toffalori C, Zito L, Gambacorta V, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25(4):603-611. doi:10.1038/s41591-019-0400-z
- Issa GC, Aldoss I, DiPersio J, et al. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature. 2023;615(7954):920-924. doi:10.1038/s41586-023-05812-3
- Candoni A, Coppola G. A 2024 Update on Menin Inhibitors. A New Class of Target Agents against KMT2A-Rearranged and NPM1-Mutated Acute Myeloid Leukemia. Hematol Rep. 2024;16(2):244-254. Published 2024 Apr 18. doi:10.3390/hematolrep16020024
- Gu Y, Nakamura T, Alder H, et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71(4):701-708. doi:10.1016/0092-8674(92)90603-a
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691-700. doi:10.1016/0092-8674(92)90602-9
- Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias [published correction appears in Proc Natl Acad Sci U S A 1992 May 1;89(9):4220]. Proc Natl Acad Sci U S A. 1991;88(23):10735-10739. doi:10.1073/pnas.88.23.10735
- Domer PH, Fakharzadeh SS, Chen CS, et al. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci U S A. 1993;90(16):7884-7888. doi:10.1073/pnas.90.16.7884
- Thirman MJ, Gill HJ, Burnett RC, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329(13):909-914. doi:10.1056/NEJM199309233291302
- Meyer C, Hofmann J, Burmeister T, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27(11):2165-2176. doi:10.1038/leu.2013.135
- Mann G, Attarbaschi A, Schrappe M, et al. Improved outcome with hematopoietic stem cell transplantation in a poor prognostic subgroup of infants with mixed-lineage-leukemia (MLL)-rearranged acute lymphoblastic leukemia: results from the Interfant-99 Study. Blood. 2010;116(15):2644-2650. doi:10.1182/blood-2010-03-273532
- Blanco JG, Dervieux T, Edick MJ, et al. Molecular emergence of acute myeloid leukemia during treatment for acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2001;98(18):10338-10343. doi:10.1073/pnas.181199898
- Chowdhury T, Brady HJ. Insights from clinical studies into the role of the MLL gene in infant and childhood leukemia. Blood Cells Mol Dis. 2008;40(2):192-199. doi:10.1016/j.bcmd.2007.07.005
- Dafflon C, Craig VJ, Méreau H, et al. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia. 2017;31(6):1269-1277. doi:10.1038/leu.2016.327
- Stein EM, Garcia-Manero G, Rizzieri DA, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131(24):2661-2669. doi:10.1182/blood-2017-12-818948
- Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275-7283. doi:10.1158/0008-5472.CAN-06-2369
- Cierpicki T, Grembecka J. Challenges and opportunities in targeting the menin-MLL interaction. Future Med Chem. 2014;6(4):447-462. doi:10.4155/fmc.13.214
- Grembecka J, He S, Shi A, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8(3):277-284. Published 2012 Jan 29. doi:10.1038/nchembio.773
- Xu Y, Yue L, Wang Y, et al. Discovery of Novel Inhibitors Targeting the Menin-Mixed Lineage Leukemia Interface Using Pharmacophore- and Docking-Based Virtual Screening. J Chem Inf Model. 2016;56(9):1847-1855. doi:10.1021/acs.jcim.6b00185
- Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30(1):41-47. doi:10.1038/ng765
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823-833. doi:10.1038/nrc2253
- Krivtsov AV, Evans K, Gadrey JY, et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell. 2019;36(6):660-673.e11. doi:10.1016/j.ccell.2019.11.001
- Wu Y, Doepner M, Hojnacki T, et al. Disruption of the menin-MLL interaction triggers menin protein degradation via ubiquitin-proteasome pathway. Am J Cancer Res. 2019;9(8):1682-1694. Published 2019 Aug 1.
- Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209-2221. doi:10.1056/NEJMoa1516192
- Falini B, Bolli N, Shan J, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107(11):4514-4523. doi:10.1182/blood-2005-11-4745
- Bolli N, Nicoletti I, De Marco MF, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67(13):6230-6237. doi:10.1158/0008-5472.CAN-07-0273
- Kühn MW, Song E, Feng Z, et al. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov. 2016;6(10):1166-1181. doi:10.1158/2159-8290.CD-16-0237
- Uckelmann HJ, Kim SM, Wong EM, et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science. 2020;367(6477):586-590. doi:10.1126/science.aax5863
- Heikamp EB, Henrich JA, Perner F, et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood. 2022;139(6):894-906. doi:10.1182/blood.2021012806
For Research Use Only. Not for Use in Diagnostic Procedures. 24-HMC-54350