When performing immunofluorescence (IF) experiments, researchers often multiplex with several antibodies and fluorescent dyes to analyze their protein of interest in relation to other cellular components. Of all the recommendation requests submitted to the IF technical support team, the most popular markers mentioned are subcellular markers.
|
|
Explore the related CST antibody sampler kit, which contains reagents to many of the markers mentioned in this blog: |
|
To help you achieve the best specificity and signal intensity in your IF experiment, this blog details the CST antibodies we recommend using to identify subcellular components depending on your required protocol.
What are subcellular markers in immunofluorescence?
Subcellular markers are high-affinity reagents designed to target organelle-specific components within a cell. These markers are invaluable for revealing the spatial location of your protein of interest and can provide a clear view of the physical and spatial characteristics of different organelles. Subcellular markers can help you study specialized organelle-specific functions, assess drug effects through organelle health, or examine cellular changes in knockout models.
When designing your experiment, you may need to select both a specific reagent for the protein of interest, as well as one or more reagents to determine subcellular localization. However, not all reagents are universally compatible. Reagents can perform differently depending on the model species and staining protocol. This guide will help you find the most suitable subcellular marker for your experiment.
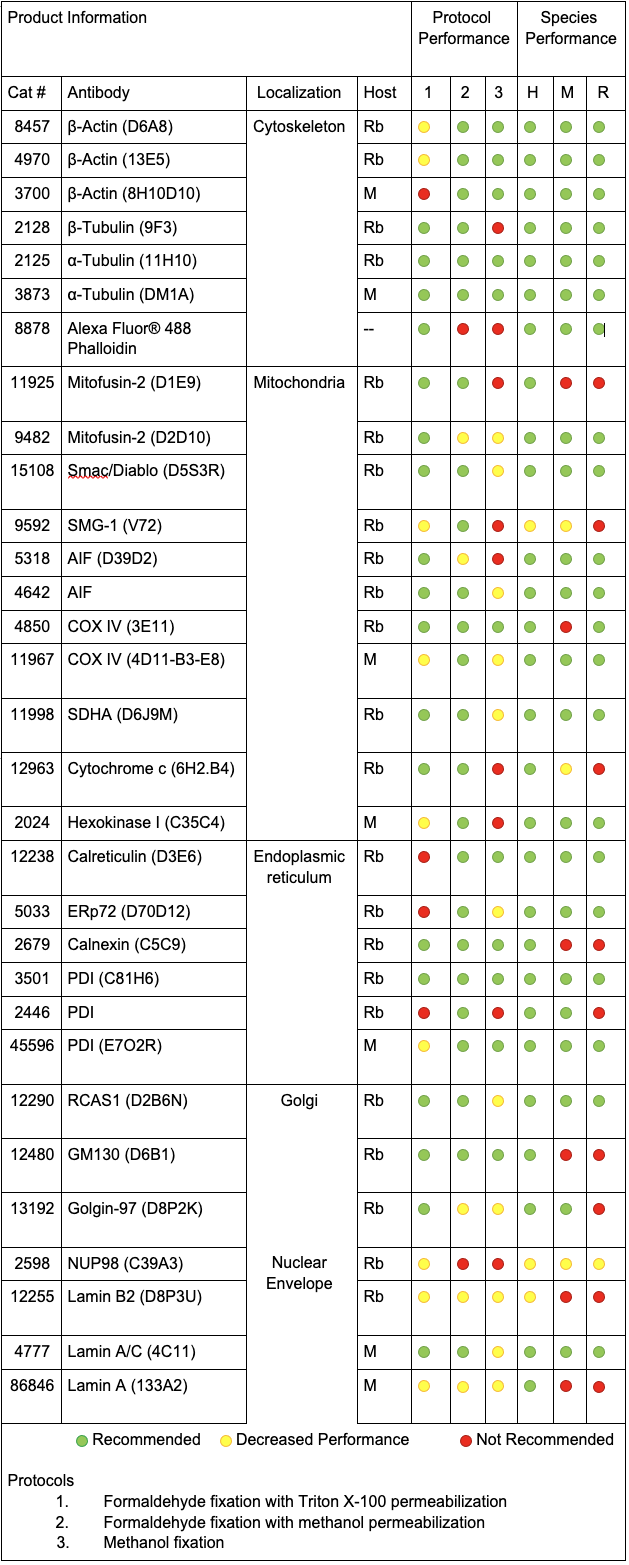
Recommended Antibodies and Protocols for Common Subcellular Markers
For the purpose of this blog, we have focused on five major subcellular localizations: cytoskeleton, mitochondria, endoplasmic reticulum (ER), Golgi apparatus, and nuclear envelope.

Note that several cellular localizations have not been included here because they either require very specific markers—for example, early endosomes (Rab5) and centrioles (PCM1)—or because faster methods are available to label them, such as the nucleus, which can be labeled using dyes like DAPI, DRAQ5, and Propidium Iodide.
%20Antibody%20%235213.webp?width=250&height=249&name=centriole%20marker_PCM-1%20(G2000)%20Antibody%20%235213.webp)
IF analysis of A204 cells using PCM-1 (G2000) Antibody #5213 (green). Actin filaments were labeled with DyLight 554 Phalloidin #13054 (red). Fluorescent DNA dye = DRAQ5® #4084. |

IF analysis of mouse retina using ProLong® Gold Antifade Reagent with DAPI #8961, Neurogranin #79519 (green), GFAP #3656 (cyan), and S6 Ribosomal Protein #5548 (red). |
The reagents we evaluated were tested in human, mouse, and rat cells using the three main protocols for we recommend for immunofluorescence experiments at Cell Signaling Technology:
- Formaldehyde fixation with Triton™ X-100 permeabilization
- Formaldehyde fixation with methanol permeabilization
- Methanol fixation
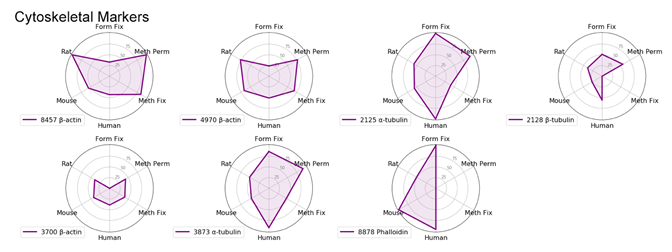
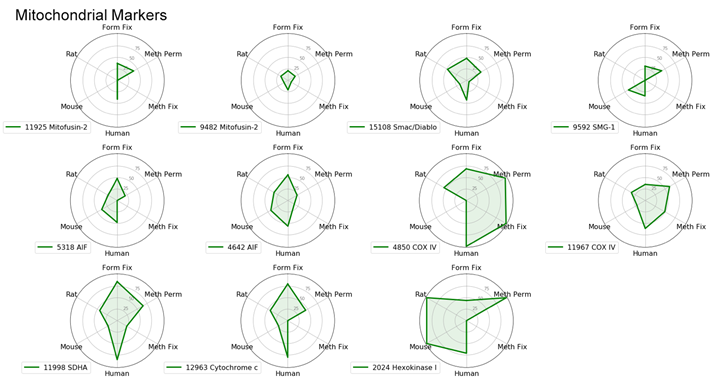
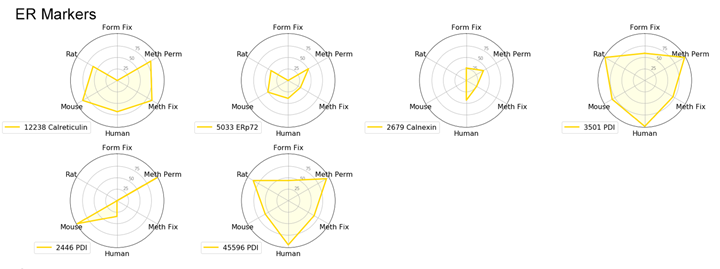
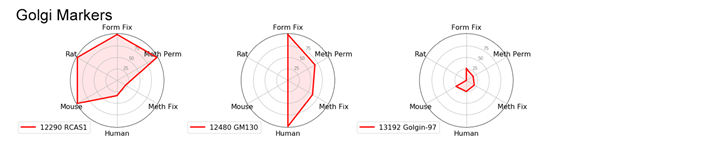
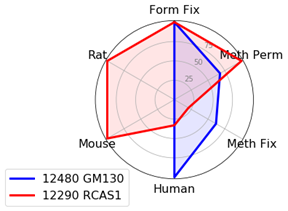
With the goal of identifying the best marker for a given protocol or species, we have compared antibodies within a given localization assessing both protocol and species performance using two criteria: specificity and signal intensity. Performance across species and protocol have been normalized to the best performer within the localization for each class and evaluated on a scale of 0 to 100. This allows us to compare multiple variables in a radar chart.
To avoid recommending products that give nonspecific patterns (e.g., nucleolar labeling with a mitochondrial marker) in a specific protocol or species, the product was given a 0 for the class it was evaluated in if nonspecific pattern was observed.
For the purpose of this experiment, each antibody was tested in triplicate in a single well of a 96-well plate diluted at the recommended dilution in 100 μL of antibody diluent. Incubation of the primary antibody was performed at 4°C overnight. Primary antibodies were labeled using host-appropriate Alexa Fluor® 488 conjugated secondary (Anti-rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) #4412 or Anti-mouse IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) #4408) antibody diluted at 1:500. Incubation of secondary antibodies were performed in the dark at room temperature for one hour. 96-well plates were analyzed by high content analysis including a laser scanning image cytometer to generate MFI data and an automated imager to generate 200X images of DAPI and the 488 channel.
You can also find a complete list of organelle marker antibodies here.
CST Recommended Antibodies and Protocols for Common Subcellular Markers
Comparing product performance throughout each subcellular localization using radar charts allowed us to identify the best performers for each category. While all radar charts are available at the end of this blog, details on the best performers for each localization are featured below.
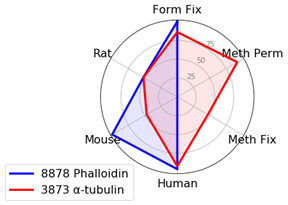
Cytoskeletal Markers: Phalloidin and α-Tubulin
When using cytoskeletal markers, the most experimental flexibility is gained by using a Phalloidin conjugate—a phallotoxin that tightly binds to filamentous actin. Because this is a non-antibody method of labeling, it allows you to conserve host species for other markers. Phalloidin has one downside for use—it is incompatible with methanol permeabilization or fixation.
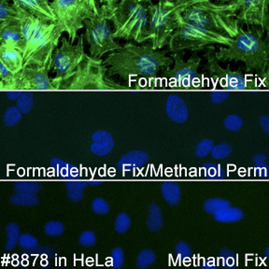
If methanol is necessary in your experiments, α-Tubulin (DM1A) Mouse mAb #3873 provides a robust signal in methanol-treated samples.
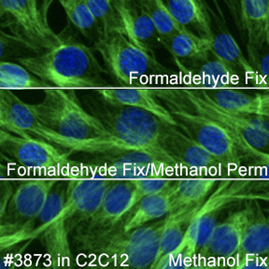
Keep in mind that tubulin labels a different component of the cytoskeleton (microtubules) compared to actin (microfilaments). The third component of the cytoskeleton—intermediate filaments—has different proteins depending on cell type, so it could not be included in this experiment.
Golgi Markers: GM130 and RCAS1
We found that golgi markers favor formaldehyde fixation with Triton™ X-100 permeabilization. However, there is substantial protocol flexibility with GM130 (D6B1) XP® Rabbit mAb #12480 if you are working with human samples.
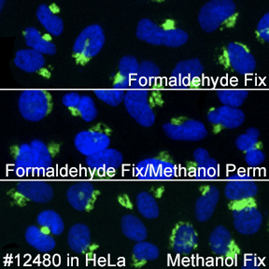
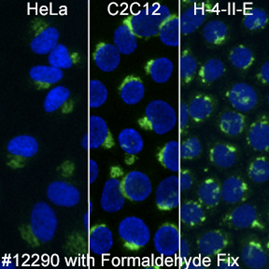
RCAS1 (D2B6N) XP® Rabbit mAb #12290 works in multiple species as long as formaldehyde fixation is included.
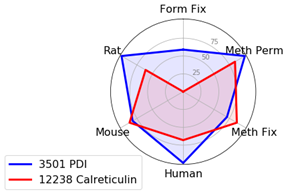
Endoplasmic Reticulum (ER) Markers: PDI or Calreticulin
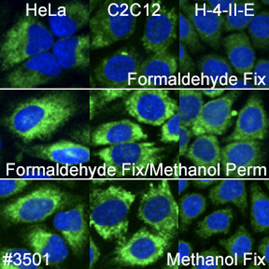
We typically recommend testing formaldehyde fixation or formaldehyde fixation with methanol permeabilization when working with ER-specific proteins. However, in this experiment, PDI (C81H6) Rabbit mAb #3501 worked well with all protocols and species tested.
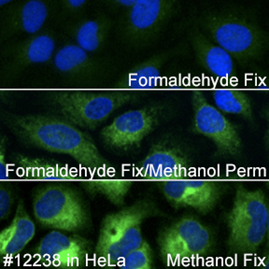
Calreticulin (D3E6) XP® Rabbit mAb #12238 is incompatible with formaldehyde fixation alone and requires either methanol permeabilization or methanol fixation.
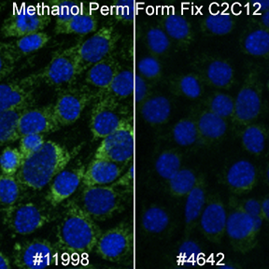
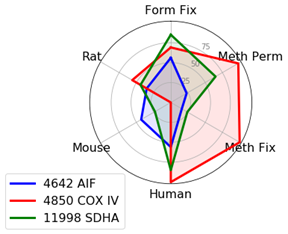
Mitochondrial Markers: AIF, COX IV, and SDHA
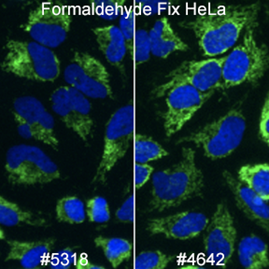
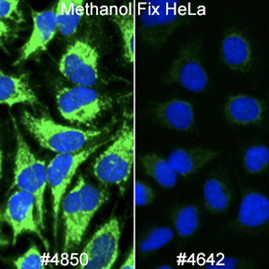
There are many available markers for mitochondria. In-house at Cell Signaling Technology, the preferred mitochondrial marker is AIF (D39D2) XP® Rabbit mAb #5318 due to compatibility with many species and protocols. AIF Antibody #4642 slightly outperforms #5318 in terms of signal; however, we prefer XP® monoclonal antibodies in-house.
In cases where methanol fixation is necessary and the species is human or rat, COX IV (3E11) Rabbit mAb #4850 may perform better than AIF antibodies.
When working with mouse cells, SDHA (D6J9M) XP® Rabbit mAb #11998 will perform slightly better than AIF antibodies in methanol permeabilized sample.
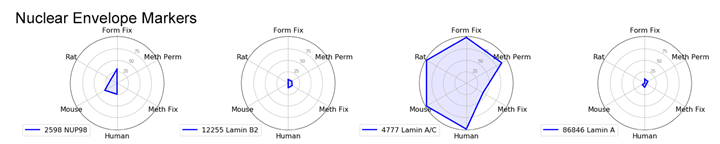
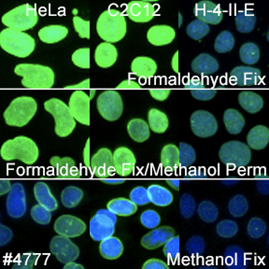
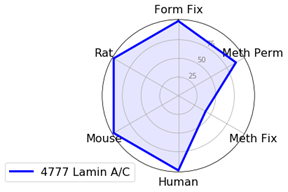
Nuclear Envelope Marker: Lamin A/C
For nuclear envelope markers, there was a clear winner. Lamin A/C (4C11) Mouse mAb #4777 worked in all species and most protocols and gave a clear, bright signal which was slightly diminished with methanol fixation.
Tips Before You Begin Your Immunofluorescence Experiment
Above we have discussed some of the best CST antibodies for each subcellular localization, depending on the protocol to be used in your experiment.
As always, the most important factor for experimental success is that samples are treated with care. Consider the health and confluence of your cells before fixation as these factors will impact your experiment. Make certain that your sample is sufficiently fixed and avoid allowing the sample to dry out, as this can negatively affect an immunofluorescence experiment.
Radar Charts