CUT&RUN delivers the power of chromatin immunoprecipitation (ChIP) or ChIP-sequencing (ChIP-seq), only takes one to two days to go from cells to DNA, and can be used with as few as 5K to 20K cells. Like any antibody-based technique, identifying a compatible antibody is a critical part of ensuring assay success. CUT&RUN is a relatively new technique, limiting the number of validated antibodies for the application. Therefore, some companies recommend using ChIP- or ChIP-seq-validated antibodies for CUT&RUN since all three techniques are used to analyze protein-DNA interactions across the genome in cells and tissues.
But do all ChIP-validated antibodies really work in a CUT&RUN assay? In fact, we have found this not to be the case. Let's explore why.
Will ChIP/ChIP-seq-validated antibodies work in a CUT&RUN assay?
Even though both ChIP and CUT&RUN assays are used to profile chromatin, there are significant differences between the methodologies that can impact antibody recognition. These differences are summarized in Table 1.
ChIP / ChIP-Seq |
CUT&RUN |
|
|
Sample Preparation |
Cross-link chromatin with formaldehyde and lyse cells. |
Bind cells to Concanavalin A-coated magnetic beads and permeabilize the cell membrane with digitonin. |
|
Chromatin Enrichment |
In vitro immunoprecipitation using an antibody specific for a DNA-binding protein or histone modification. |
In vivo enrichment, where the antibody specific for a DNA-binding protein, cofactor, or histone modification enters the nuclei after cell membrane permeabilization. |
|
Chromatin Fragmentation |
Non-targeted enzymatic digestion with MNase or mechanically using sonication to shear the chromatin prior to chromatin immunoprecipitation. |
Targeted enzymatic digestion during enrichment based on the interaction of a primary antibody with pAG-MNase. |
|
Chromatin/Protein State |
Fragmented chromatin containing partially denatured histones and DNA-binding proteins. |
Contiguous chromatin with histones and DNA-binding proteins in their native conformations. |
Table 1: ChIP and CUT&RUN methodologies.
Not all ChIP- and ChIP-seq-Validated Antibodies Work in CUT&RUN
We tested antibodies for over 100 (and counting!) target proteins and found that only 50-60% of ChIP- or ChIP-seq antibodies were compatible with CUT&RUN.
ChIP experiments were run using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads) #9005, while CUT&RUN experiments were performed using the CUT&RUN Assay Kit #86652 and DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN) #14209. Downstream quantitative polymerase chain reaction (qPCR) analyses were performed using SimpleChIP Universal qPCR Master Mix #88989, and DNA library preparation was done using the DNA Library Prep Kit for Illumina (ChIP-seq, CUT&RUN) #56795 and Multiplex Oligos for Illumina (Dual Index Primers) #47538. NG-seq was performed using an Illumina NextSeq Platform.
Stat1 Antibody Testing
When Stat1 (D1K9Y) Rabbit mAb #14994, which is ChIP-validated, and Stat1 (D4Y6Z) Rabbit mAb #14995, which is ChIP- and ChIP-seq-validated, were tested, both antibodies demonstrated robust enrichment of target genes in a ChIP assay (Figure 1A). Stat1 #14994 did not generate passable data for ChIP-seq, but it did show enrichment in the CUT&RUN assay. On the other hand, Stat1 #14995, which is validated for ChIP and ChIP-seq, did not work for CUT&RUN since there was no observed enrichment for known STAT1 target genes IRF1 or AIM2 (Figure 1B).
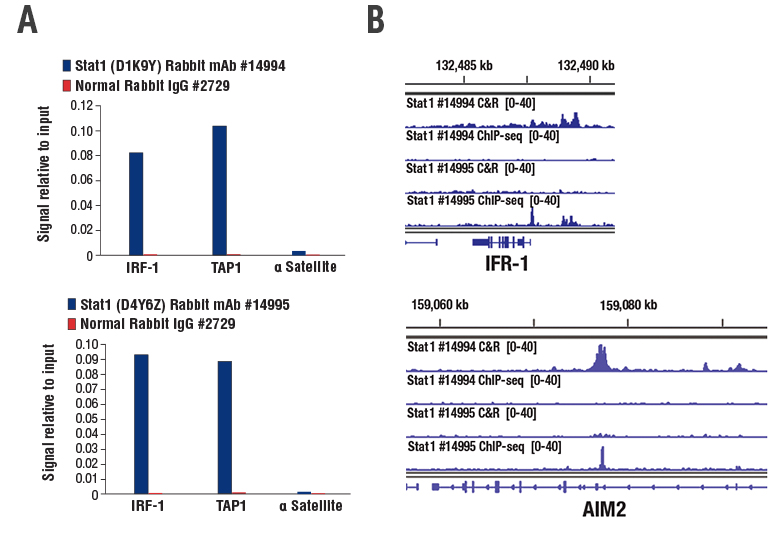 Figure 1. ChIP and CUT&RUN experiments were performed on HT-1080 cells treated with Human Interferon-γ (hIFN-γ). While both STAT1 rabbit monoclonal antibodies show robust enrichment of target genes in ChIP (A), only Stat1 (D1K9Y) Rabbit mAb #14994 shows target gene enrichment in CUT&RUN (B). Note that while the Stat1 (D4Y6Z) Rabbit mAb #14995 is validated for and shows target gene enrichment in ChIP-seq, it does not work for CUT&RUN (B).
Figure 1. ChIP and CUT&RUN experiments were performed on HT-1080 cells treated with Human Interferon-γ (hIFN-γ). While both STAT1 rabbit monoclonal antibodies show robust enrichment of target genes in ChIP (A), only Stat1 (D1K9Y) Rabbit mAb #14994 shows target gene enrichment in CUT&RUN (B). Note that while the Stat1 (D4Y6Z) Rabbit mAb #14995 is validated for and shows target gene enrichment in ChIP-seq, it does not work for CUT&RUN (B).
Stat3 Antibody Testing
Three Stat3 antibodies were also tested: Stat3 (124H6) Mouse mAb #9139 and Stat3 (79D7) Rabbit mAb #4904, which are both ChIP- and ChIP-seq-validated, and Stat3 (D3Z2G) Rabbit mAb #12640. Again, all three antibodies robustly enriched target genes using ChIP (Figure 2A), but only Stat3 #9139 demonstrated robust target gene enrichment with CUT&RUN (Figure 2B). Stat3 #12640 did show some target gene enrichment in CUT&RUN, but ultimately did not meet Cell Signaling Technology (CST) validation criteria because a low genomic signal-to-noise ratio was observed.
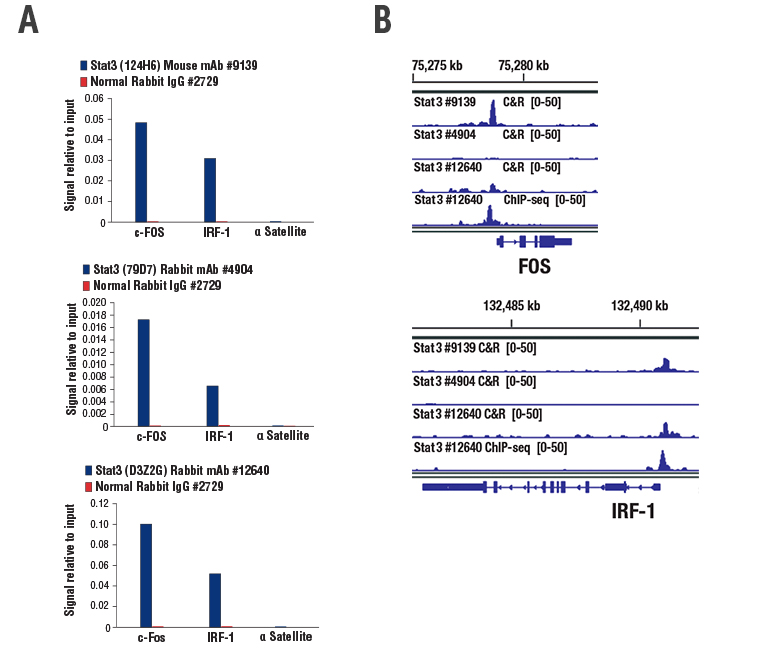 Figure 2. ChIP and CUT&RUN experiments were performed with HepG2 cells starved overnight and treated with IL-60. While all three STAT3 monoclonal antibodies show robust enrichment of target genes in ChIP (A), only Stat3 (124H6) Mouse mAb #9139 shows robust target gene enrichment in CUT&RUN (B). Note that while the ChIP-seq-validated Stat3 (D3Z2G) Rabbit mAb #12640 does show enrichment of target genes in CUT&RUN (B), it failed validation due to high background signal.
Figure 2. ChIP and CUT&RUN experiments were performed with HepG2 cells starved overnight and treated with IL-60. While all three STAT3 monoclonal antibodies show robust enrichment of target genes in ChIP (A), only Stat3 (124H6) Mouse mAb #9139 shows robust target gene enrichment in CUT&RUN (B). Note that while the ChIP-seq-validated Stat3 (D3Z2G) Rabbit mAb #12640 does show enrichment of target genes in CUT&RUN (B), it failed validation due to high background signal.
Antibody |
ChIP-Validated |
ChIP-seq Validated |
Works on CUT&RUN |
|
Stat1 (D1K9Y) Rabbit mAb #14994 |
Yes |
No |
Yes |
|
Stat1 (D4Y6Z) Rabbit mAb #14995 |
Yes |
Yes |
No |
|
Stat3 (124H6) Mouse mAb #9139 |
Yes |
No |
Yes |
|
Stat3 (79D7) Rabbit mAb #4904 |
Yes |
No |
No |
|
Stat3 (D3Z2G) Rabbit mAb #12640 |
Yes |
Yes |
No |
Table 2: Summary of described antibodies tested on CUT&RUN, a small subset of the total antibodies tested by CST.
As these two examples illustrate, relying solely on the fact that an antibody is validated for ChIP or ChIP-seq does not necessarily guarantee success in a CUT&RUN assay. It's best to use a CUT&RUN-validated antibody to ensure success. However, you can start with ChIP- or ChIP-seq-validated antibodies if a CUT&RUN-validated antibody hasn’t been identified yet. Just be aware that you’ll likely need to screen different antibodies to find one that will work.
To save you time and money you’d otherwise spend testing antibodies yourself, CST is actively validating CUT&RUN antibodies for you. Browse our catalog of over 150 CUT&RUN-validated antibodies:
How CST Validates an Antibody for CUT&RUN
CST is known for having stringent validation standards to ensure antibodies will work for your application, and it’s no different when it comes to validating antibodies for CUT&RUN. To provide antibodies that are proven to work, we test all of our antibodies using the CUT&RUN Assay Kit #86652, the DNA Purification Buffers and Spin Columns (ChIP, CUT&RUN) #14209, the SimpleChIP Universal qPCR Master Mix #8898, the DNA Library Prep Kit for Illumina #56795 (ChIP-seq, CUT&RUN), and the Multiplex Oligos for Illumna (Dual Index Primers) #47538.
Antibodies must pass all of the following criteria to qualify as a CUTRUN-validated reagent:
- Antibody Sensitivity: Compare the target enrichment signal-to-noise ratio to input controls. The antibody must provide an acceptable minimum number of defined enrichment peaks and a minimum signal-to-noise ratio compared to input chromatin to pass.
- Antibody Specificity:
- Compare the signal from knock-out cell lines with wild-type cells. If knock-out cells are not available, look for a consistent profile of enrichment peaks across the genome using multiple antibodies against distinct target protein epitopes.
- Look for a consistent profile of enrichment peaks using antibodies raised against different subunits in a multiprotein complex.
- Compare enrichments across the genome to published CUT&RUN and ChIP-seq data (i.e., ENCODE) using additional antibodies for a given target protein.
- Analyze enrichment of known positive and negative target loci using qualitative PCR (qPCR). The antibody must show a minimum fold enrichment for the known positive loci compared to the known negative target loci. It also must show a minimum signal-to-noise ratio determined by the level of enrichment of the known target loci by the target-specific antibody vs. an isotype control antibody.
- Antibody specificity for sequence-specific DNA-binding transcription factors: Perform transcription factor binding-motif analysis of enriched chromatin fragments.
Using ChIP- or ChIP-seq-validated antibodies for CUT&RUN does not automatically guarantee success since only 50-60% of them are compatible. Before you spend time screening antibodies yourself, be sure to check our ever-growing list of CUT&RUN antibodies validated by CST. You may be able to save yourself valuable time!
Additional Resources
- Blog: ChIP, CUT&RUN & CUT&Tag: Support Reagents for Successful Chromatin Profiling
- Don’t see a validated antibody for your target of interest? Contact Tech Support, as we may be in the process of testing your antibody of interest.
- Need more info? Check out the CUT&RUN Resource Center, where you can find a list of validated antibodies, videos, troubleshooting guides, and FAQs, including the following:







