For decades, immunohistochemistry (IHC) has been a powerful technique for the investigation and visualization of cellular components in their native histological context. IHC has served as an important tool in medicine – enabling the diagnosis of complex pathological conditions – and in basic research to advance the understanding of key biological processes.
IHC protocols involve many steps – from tissue processing to signal detection – that require significant time and the expertise of trained technicians to complete by hand. In recent years, the task of IHC has been dramatically improved by the development of automated methods. Autostainers, like the Leica Bond, enable increased workload throughput while also providing increased consistency and reproducibility of staining. However, like manual staining, automated IHC requires the tailoring of each protocol to suit the specific needs of each experiment.
In a separate blog post, we provided an overview of the basic principles of IHC and guidelines for optimizing the manual detection of two targets of interest using a dual chromogenic stain, steps that can be readily adapted for use on an autostainer. A critical recommendation in that procedure was to select validated antibodies raised in different host species to detect individual target antigens. Doing so, limits the possibility of cross-reactivity during signaling processing. However, the luxury of having validated primary antibodies from different host species is not always an option. In that case, it is necessary to establish best practices for dual staining with primary antibodies raised in the same host species.
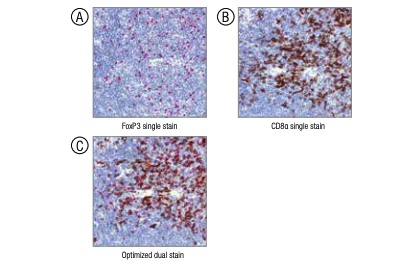 Comparison of the single-stain images of (A) FoxP3 (#12653) and (B) CD8α (#98941) to the optimized dual stain (C).
Comparison of the single-stain images of (A) FoxP3 (#12653) and (B) CD8α (#98941) to the optimized dual stain (C).
What are the steps necessary to optimize the sequential detection of two antigens by IHC with primary antibodies raised in the same species? As was the case when using primary antibodies from different host species, the first step is to perform control stains for individual primary antibody/chromogen pairs to establish a baseline for the detection of each antigen. Due to differences in binding affinity and reactivity, it is important to mix and match the primary antibodies and chromogen-tagged secondary antibodies to identify the best combination.
Next, it is essential to develop and test an antibody stripping step to remove residual primary antibody following staining for the first antigen to limit cross-reactivity during the detection of the second antigen. Efficient stripping can be determined in the following manner: Perform tissue incubation with the primary antibody to antigen #1, remove the antibody with stripping buffer, and then add detection reagents for antigen #2. Any signal detected is indicative of the presence of residual primary antibody that could compromise the interpretation of the results following the sequential detection of the antigen #2.
Stripping optimization should be performed for each primary antibody/chromogen pair. Pairings that show the most effective stripping will determine the order of operations in the sequential stain, i.e. the antibody/chromogen pair that shows little to no staining in the stripping test should be used first in the dual stain. Following these steps ensures confidence in the final interpretation of dual staining with primary antibodies derived from the same host species.
For more details on this procedure, we encourage you to refer to our recent application note: Optimization of mouse CD8 and FoxP3 dual staining on the Leica® Bond™.