What if you could measure RNA, determine cell type, and detect intracellular signaling, all in a single cell? Theoretically, such an assay would be a Rosetta Stone of signaling research, capable of changing our understanding of disease progression and therapeutic response. This comprehensive dataset would allow us to directly measure how cell type affects RNA level and protein expression and vice versa, enabling more in-depth insights into the differences between diseased and healthy tissues.
For years, scientists have been attempting to make such an assay a reality, but have struggled to measure intracellular data and post-translational modifications (PTMs) without degrading RNA. Novel methodologies have come close, but often lack reproducibility across industries, laboratories, and applications.
Now, a new method that leverages cutting-edge 10x Genomics Chromium gene expression technology with Feature Barcode and stringently validated antibodies from CST promises to do just that: The InTraSeq™ Single Cell Analysis assay.
 Figure 1. Left: Most single cell analysis techniques detect RNA and use barcoded antibodies to measure surface proteins for phenotyping purposes. Center: Many methods developed to detect intracellular proteins suffer from significant RNA loss and degradation. Right: The InTraSeq assay detects RNA alongside intracellular and surface proteins while preserving RNA.
Figure 1. Left: Most single cell analysis techniques detect RNA and use barcoded antibodies to measure surface proteins for phenotyping purposes. Center: Many methods developed to detect intracellular proteins suffer from significant RNA loss and degradation. Right: The InTraSeq assay detects RNA alongside intracellular and surface proteins while preserving RNA.
"InTraSeq is a technology whereby we can, in fact, look at the single level at the signaling components within the cell and combine it together with scRNA-seq," explains Vijay Kuchroo, PhD, DVM, Samuel L. Wasserstrom Professor of Neurology at Harvard Medical School and Senior Scientist at Brigham and Women's Hospital. "It's actually going to be a revolutionary technology."
But why has it been so challenging to capture both single cell RNA and intracellular signaling data in a single sample? To appreciate the breakthrough, it’s important to understand the challenges researchers face in combining transcriptomic and proteomic information.
Developing a Reproducible Multi-Omic & Multimodal scRNAseq Assay
Historically, cellular signaling research has been conducted at the population level—researchers grind up cells and use techniques like western blotting or immunoprecipitation to analyze PTMs. However, this approach only provides an average view of signaling activity across an entire cellular population.
“We know from single cell RNA sequencing that individual cells within a heterogenous sample have functions that are very different from one another,” explains Majd Ariss, Senior Scientist at CST who was closely involved in developing the InTraSeq technology. “This distinction is especially important in diseased tissues, such as complex tumors and their microenvironment, where each different cell type demonstrates distinct biological processes depending on their spatial location.”
The main challenge so far has been the ability to simultaneously obtain transcriptomics data from scRNA-seq experiments, as well as functional readouts of cell type from assays that identify intracellular PTMs and/or cytoplasmic proteins. Various methodologies can achieve one or even two of these analyses in isolation, but not all of them simultaneously within a single sample, therefore providing only a partial view of cellular biology.
"RNA is prone to degradation, and most methods that attempt to detect intracellular proteins end up destroying RNA in the process," explains Ariss. "This has made it extremely difficult to obtain both transcriptomic and proteomic information from the same cell."
Additionally, the timescales of signaling events and transcriptional changes are vastly different, and bridging the temporal gap to understand how rapid signaling events lead to longer-term transcriptional changes has been a significant challenge (Figure 2).
 Figure 2. Signaling events happen in minutes to hours, while RNA transcription and the translation of proteins takes ~1 hour to days.
Figure 2. Signaling events happen in minutes to hours, while RNA transcription and the translation of proteins takes ~1 hour to days.
"Signaling happens within minutes to an hour, but transcription happens in days—hours and days," explains Kuchroo. "So, to combine what happened in 10 minutes and what impact it has on the days, and the function later, is actually an interesting problem that needed to be solved. And I think InTraSeq is going to be addressing that problem."
Ariss and the team at CST worked with Dr Kuchroo as well as other leading researchers to develop computational methods that accurately reconcile the differing timescales of signaling, transcription, and translation.
"We were closely involved with Majd at CST to brainstorm what they came up with, and how to use those tools to address the problem," says Kuchroo. This included "not only running some of the InTraSeq antibodies and components, but also, computationally, how to combine [the data across different timescales]."
The result is the InTraSeq technology, a multimodal scRNA-seq assay that leverages oligo-barcoded antibodies to enable RNA and protein profilings—including intracellular PTMs—from a single sample using a relatively simple protocol. This breakthrough allows researchers to directly link signaling events to transcriptional outputs within individual cells, providing a more comprehensive understanding of cellular heterogeneity and function.
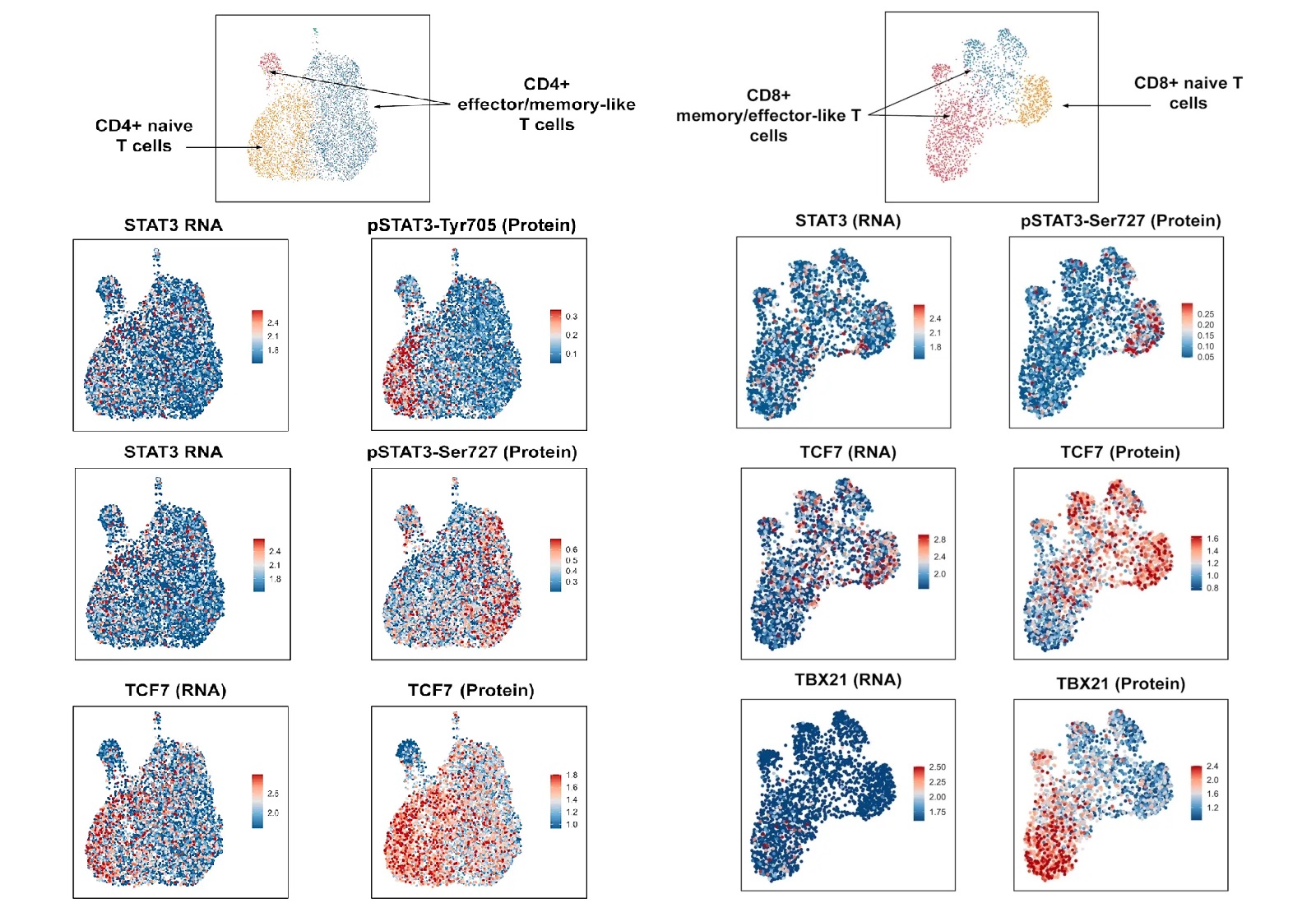 Figure 3. Analysis of isolated CD4+ and CD8+ cells using InTraSeq intracellular and signaling pathway antibodies to identify cellular states that would be difficult to study using RNA-seq alone. The InTraSeq assay provides unparalleled insights into cellular subpopulations, enabling a deeper understanding of disease mechanisms.
Figure 3. Analysis of isolated CD4+ and CD8+ cells using InTraSeq intracellular and signaling pathway antibodies to identify cellular states that would be difficult to study using RNA-seq alone. The InTraSeq assay provides unparalleled insights into cellular subpopulations, enabling a deeper understanding of disease mechanisms.
"Now, computationally, not only are you looking at the molecules that signal, you are putting together the signaling network inside the cell, and then looking at... what the signaling pathway is actually inducing at the transcriptional level," explains Kuchroo.
Insights from Bridging RNA and Protein Data
It has long been assumed that the presence of RNA correlates with protein synthesis, and that the abundance of mRNA can be used as a proxy to infer protein levels. However, multimodal scRNA-seq assays like the InTraSeq technology have shown that this may not always be the case. RNA degradation and the different timelines between transcription and translation can create discrepancies in the data gathered using traditional assays.
"Using the InTraSeq technology, we've found that RNA levels can differ from protein expression," explains Ariss. "This early research underscores the importance of studying these processes together in the same cell to fully understand how signaling pathways influence transcription and ultimately inform cellular function."
Ariss' research, published in the preprint paper InTraSeq: A Multimodal Assay that Uncovers New Single Cell Biology and Regulatory Mechanisms, was conducted in collaboration with scientists including Dr Kuchroo at The Gene Lay Institute of Immunology and Inflammation of Brigham and Women’s Hospital, Massachusetts General Hospital, and Harvard Medical School.
By bridging the gap between RNA transcription and protein activity, the InTraSeq assay offers a more nuanced understanding of cellular biology. This has the potential to uncover previously hidden cellular states and mechanisms, deepening our understanding of disease progression and therapeutic response.







