We’re proud to partner with The Michael J. Fox Foundation for Parkinson’s Research (MJFF) to move PD research forward. Learn more about the partnership and explore PD scientific resources.
—
The number of people affected by Parkinson’s disease is growing, with more than eight million of those over fifty predicted to have PD by 2030.1 The disease is characterized by the progressive loss of dopaminergic neurons in the midbrain resulting from impairments in multiple cellular pathways, and there currently is no cure. Developing effective treatments for PD demands high-quality antibodies to investigate the underlying mechanisms of neurodegeneration.
The Leucine-rich repeat kinase 2 (LRRK2) gene is the most common genetic cause of PD, and it is thought that mutations in LRRK2 contribute to disease progression by impacting the kinetics of the autophagic-lysosomal pathway (ALP) in neurons.2 Patient-derived induced pluripotent stem cells (iPSCs) represent powerful tools for studying the impact of LRRK2 mutations and generating novel antibody reagents.
Download the Neurodegeneration Signaling Pathways
With support from FUJIFILM Cellular Dynamics, my colleagues and I developed a portfolio of rabbit and mouse monoclonal antibodies for organelle and central nervous system (CNS) markers to help with assessing cellular processes in PD. Our antibody validation strategy included the use of high-throughput imaging for quantifying endosome and lysosome protein expression in iPSC-derived dopaminergic neurons (iCell® DopaNeurons) generated from:
- An apparently healthy normal donor
- A PD-diagnosed donor harboring a LRRK2 G2019S mutation
- A PD-diagnosed donor with a mutation-corrected LRRK2
Read on to learn more about how we validated our antibodies for specific labeling of ALP-associated proteins in these in vitro models. At the end of this post, you’ll also find a list of rabbit and mouse monoclonal antibodies we developed for organelle and CNS markers.
Identification of iPSC-derived Dopaminergic Neurons
At Cell Signaling Technology (CST), we adhere to the Hallmarks of Antibody Validation™ to ensure that our antibodies are highly specific and perform as expected in their intended application. By carefully tailoring which of these six complementary validation strategies is applied to each antibody, we guarantee that our antibodies are fit for purpose based on factors including the availability of appropriate testing models and the biological role of the target.
When developing our portfolio of antibodies for PD research, we collaborated with FUJIFILM Cellular Dynamics to leverage their human iPSC-derived dopaminergic neurons (iCell® DopaNeurons: Apparently Healthy Normal - #R1032, LRRK2 G2019S - #R1234, LRRK2 G2019S/G mutation-corrected control - #R1243) for validation purposes. Because these neurons are commercially available and fully differentiated upon thaw, they allow for the rapid generation of biologically relevant and reproducible results.
The iPSC-derived dopaminergic neurons were cultured following the iCell DopaNeurons User’s Guide. After 14 days in culture, the cells were fixed in methanol and stained for various cellular markers and CNS cell types utilizing either the Immunofluorescence Protocol with Formaldehyde Fixation or the Immunofluorescence Protocol for Cell-based Assays Requiring Methanol Fixation. Images were captured at 63X using the Operetta CLS HCA system in confocal mode, prior to quantification with Harmony high-content analysis software, Mitochondria Analyzer for ImageJ,3 and CellProfiler.4
Figure 1 shows representative immunofluorescent staining of control and LRRK2 G2019S iPSC-derived dopaminergic neurons using CST antibodies for Neuronal nuclei (NeuN), Tyrosine hydroxylase (TH), β3-Tubulin, and Forkhead box protein A2 (FoxA2), confirming the dopaminergic nature of the neurons.
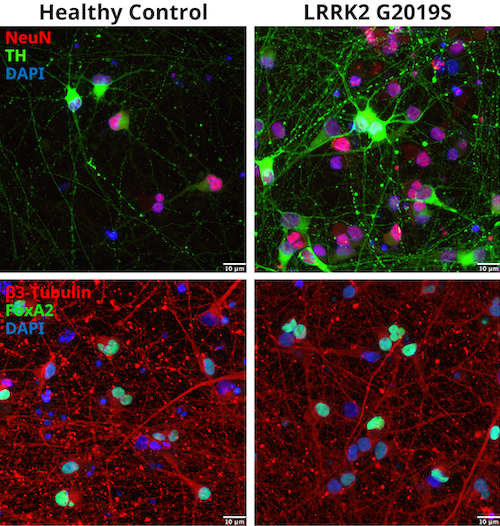
Figure 1: Identification of iCell® DopaNeurons. Donor-derived dopaminergic healthy, WT neurons and cells harboring a LRRK2 G2019S mutation were stained with NeuN (E4M5P) Mouse mAb #94403 and Tyrosine Hydroxylase (E2L6M) Rabbit mAb #58844 (upper panel) or β3-Tubulin (E9F3E) Mouse mAb #45058 and FoxA2/HNF3β (D56D6) XP® Rabbit mAb #8186 (lower panel).
Assessment of Lysosomes in iCell® DopaNeurons with LRRK2 Mutation
Lysosomes were assessed by staining for Lysosome-associated membrane protein 1 (LAMP1) and Cathepsin B (CTSB) in all three types of iPSC-derived dopaminergic neurons, using β3-Tubulin and DAPI for counterstaining. The mean fluorescent intensity (MFI), spots per neuron, and spot area were then quantified, as shown in Figure 2.
Figure 2: Lysosomal proteins imaged in iCell® DopaNeurons. Lysosomal proteins LAMP1 (D2D11) XP® Rabbit mAb #9091 (A) and Cathepsin B (D1C7Y) XP® Rabbit mAb #31718 (E) were imaged in healthy control iCell® DopaNeurons, neurons harboring a LRRK2 G2019S mutation, and LRRK2 mutation-corrected control (MCC) neurons. The images were quantified for mean fluorescent intensity (B, F), spots per neuron (C, G), and spot area (D, H).
The MFI for both LAMP1 and Cathepsin B was trending lower in the LRRK2 G2019S neurons as compared to the healthy control and mutation-corrected neurons, which is to be expected since lysosomal deficiency is a characteristic of PD. When we looked at lysosomal number and size by respectively quantifying LAMP1+ spots per neuron and the spot area, no differences between genotypes were observed.
Neurons with the LRRK2 G2019S Mutation Display Increased LC3 Signal
Autophagy was compared across the three different cell types by staining for Light Chain 3 (LC3) using LC3A/B (D3U4C) XP® Rabbit mAb #12741. Figure 3 shows that LC3 is increased in the LRRK2 G2019S neurons and rescued to normal levels in the LRRK2 mutation-corrected control (MCC) cells.
Figure 3: Quantification of LC3 staining. iCell® DopaNeurons were stained with LC3A/B (D3U4C) XP® Rabbit mAb #12741 (A) and the mean fluorescent intensity was quantified (B).
LRRK2 G2019S Does Not Change Mitochondria Morphology
To determine whether mitochondrial number and gross morphology were affected by the LRRK2 G2019S mutation, healthy and LRRK2 G2019S neurons were stained for Cytochrome c oxidase IV (COX IV), a widely used mitochondrial-marker protein. No significant differences were observed between the two cell types, as shown in Figure 4.
Figure 4: Mitochondria morphology assessment in iPSC-derived neurons. iCell® DopaNeurons were stained with COX IV (3E11) Rabbit mAb #4850 (A) and a threshold was applied (B) to quantify mitochondrial parameters. No change was found in the number of mitochondria per neuron (C), branches per mitochondria (D), and branch junctions per neuron (E).
The Future of PD Research
With no cure currently available for PD, researchers require high-quality antibodies to advance their understanding of this prevalent neurodegenerative disorder. We have developed a portfolio of rabbit and mouse monoclonal antibodies for organelle and CNS markers that can help assess cellular processes in PD, using commercially available human iPSC-derived dopaminergic neurons to support our rigorous validation process.
Our data suggest that PD donor-derived dopaminergic neurons harboring a LRRK2 G2019S mutation exhibit elevated LC3 expression, which is rescued in neurons corrected for the LRRK2 mutation, and that the LRRK2 G2019S mutation has no impact on the number and gross morphology of lysosomes and mitochondria.
|
|
View the poster from SfN, Characterization of the autophagic-lysosomal pathway in Parkinson’s disease using patient iPSC-derived dopaminergic neurons containing a LRRK2 G2019S mutation. |
|
We continue to develop further antibodies to facilitate PD research, tailoring our validation strategies to ensure high specificity and reliable performance.
Monoclonal Antibodies for Organelle and CNS Markers
Select References:
- Dorsey ER, Constantinescu R, Thompson JP, et al. Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology. 2007;68(5):384-386
- Madureira M, Connor-Robson N, Wade-Martins R. "LRRK2: Autophagy and Lysosomal Activity". Front Neurosci. 2020 May 25;14:498. doi: 10.3389/fnins.2020.00498. PMID: 32523507; PMCID: PMC7262160.
- Chaudhry A, Shi R, Luciani DS. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am J Physiol Endocrinol Metab. 2020;318(2):E87-E101. doi:10.1152/ajpendo.00457.2019
- Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics. 2021;22(1):433. Published 2021 Sep 10. doi:10.1186/s12859-021-04344-9
Alexandra Foley, Scientific Marketing Writer and CST Blog Manager, contributed to writing this post.24-NDT-69935
