Understanding how antibodies are made and the pros and cons of the different types of antibodies is critical when it's time to design your experiments and generate reproducible data. In Part 1 of the Antibody Essentials series, we discussed how antibodies are made, their structure and function, and other antibody basics. Here, we’ll take a look at the biological mechanisms that allow for the incredible functional diversity of antibodies and consider the various ways that antibodies can be classified.
Table of Contents
Understanding Antibody Diversity and Classification
As you already know, an antibody is a complex protein made by a particular class of immune cells, called B cells, during the adaptive immune response. The immune system needs the ability to recognize thousands of different potential pathogens that it may encounter over the lifetime of the animal, but with an almost infinite number of antigens available, how can B cells produce antibodies against all these potential targets? To address this question, let’s briefly explore some of the basics of how B cells develop to understand their role in immunology.
How is antibody diversity generated?
Following maturation in the bone marrow and spleen, every circulating B cell expresses a unique B cell receptor (BCR), composed of a membrane immunoglobulin (antibody) and associated Igα/Igβ (CD79a/CD79b) heterodimers. When the BCR encounters and recognizes an antigen, the B cell internalizes and processes the antigen. A fragment of the antigen is bound to a major histocompatibility complex (MHC) class II molecule to form a complex, and this fragment-MHC class II complex is ultimately displayed on the B cell surface. Receptors on T helper (Th) cells recognize the complex and bind it, activating the Th cell, which in turn produces cytokines that activate the B cell to become a proliferative B plasma cell. This leads to rapid clonal expansion of a population of identical, antibody-secreting plasma cells that recruit immune cells to kill the pathogen and promote antigen clearance by the immune system. Although many plasma cells are only short-lived, a proportion survive to become memory B cells that perform a protective function upon subsequent exposure to the same antigen.
The effectiveness of adaptive immunity hinges on the fact that each B cell clone expresses a different antibody at its surface. So, where does that come from?
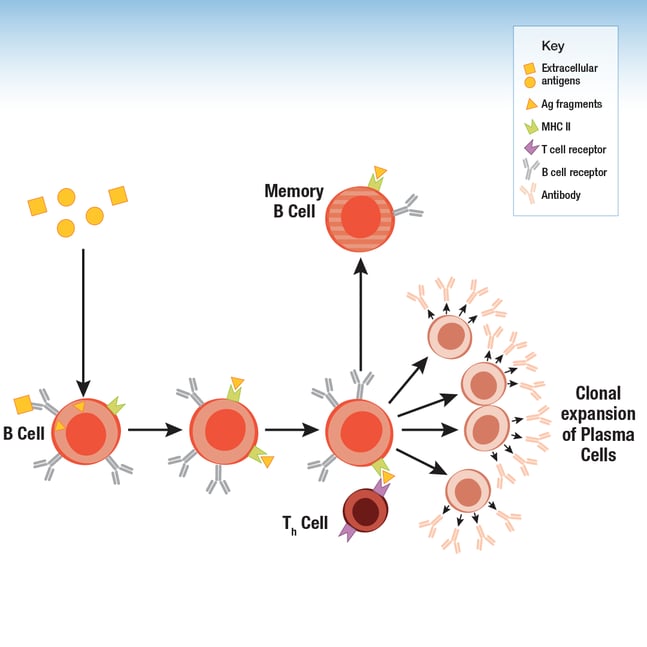 Figure 1. B cells display antigen fragments via the MHC II complex on their surface. During an immune response, Th cells stimulate clonal expansion of B cells to drive increased production of the antibody.
Figure 1. B cells display antigen fragments via the MHC II complex on their surface. During an immune response, Th cells stimulate clonal expansion of B cells to drive increased production of the antibody.
Antibody diversity at the B cell surface is achieved early in B cell development, prior to maturation, through a process that includes DNA rearrangement of the genes that code for the antibody heavy and light chains. This occurs via three major mechanisms: V(D)J recombination, junctional diversification, and somatic hypermutation (Figure 2).
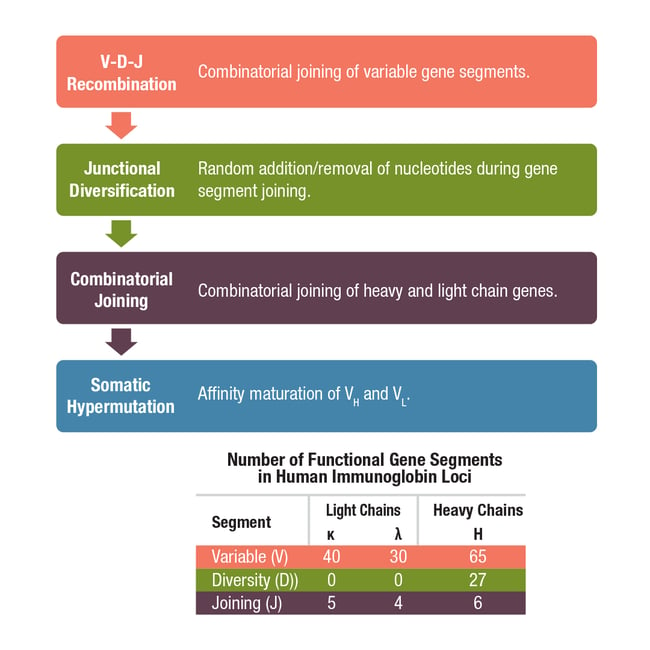
Figure 2. Mechanisms involved in the generation of antibody diversity.
First let’s consider V(D)J recombination, which is the splicing of different variable, diversity, and joining gene segments during B cell development. Each antibody heavy chain results from a single constant chain gene being spliced to one variable gene segment, one diversity gene segment, and one joining gene segment, whereas light chains are composed of V (kappa/κ or lambda/λ) and J segments, but do not include a D segment. The sheer number of V, D, and J gene segments available (Figure 2), along with the random recombination of segments by lymphocyte-specific recombinase complexes, results in millions of possible genetic configurations. This diversity is further increased by junctional diversification, whereby random nucleotides are lost from or added at the ends of V-region coding sequences as they are joined by the V(D)J enzyme complex.
Once V(D)J recombination and junctional diversification are complete, the heavy and light chain gene sequences are arranged in the correct order to produce functional antibody polypeptides. However, the recombined genes are subject to a third level of diversification called somatic hypermutation. Somatic hypermutation involves the accumulation of point mutations in the variable region of the heavy and light chains and occurs following B cell activation. The rate of these mutations is about a million times higher than the background mutation rate for other genes to further diversify the pool of antibodies. This results in higher-affinity antibodies as the adaptive immune response progresses, a process referred to as affinity maturation. Together, these diversification mechanisms allow for the production of antibodies capable of responding to billions of unique antigens, enough to ensure an immune response to any non-self antigen.
Further Reading: Janeway CA Jr, Travers P, Walport M, et al.The generation of diversity in immunoglobulins. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001.
How antibodies are classified?
Now that we’ve considered the mechanisms behind antibody diversity, it’s time to think about antibody classification. Antibodies are commonly classified in numerous ways, including:
- Target reactivity
- Species reactivity
- Host species used for immunization/antibody production
- Polyclonal or monoclonal
- Research use only, diagnostic use, or therapeutic use
- Recombinant or non-recombinant antibody production
Below, we'll explore each of these antibody classification categories in more detail.
Target Reactivity
Target reactivity is likely the first thing you think about when you begin to select antibodies to use in your experiment. A well-characterized antibody that is both specific and selective will react with and bind to its target biomolecule (most commonly a protein), and will not react with off-target molecules. The name of the target protein is usually included in the name of the antibody product to indicate the expected target reactivity.
For many targets, there will be more than one antibody available that reacts with the target, which could recognize different epitopes of the same target and have slightly different performance characteristics. So, it's important to dig into the validation data and learn more about each one when designing your experiments.
Antibodies that are specific but less selective may still be valuable tools when used appropriately. An example is this pan-Cadherin antibody, which reacts with a conserved antigen expressed in multiple members of the Cadherin family. A screening experiment with a pan-specific antibody could be followed up with more selective antibodies to zero in on individual targets within that family.
Many proteins undergo post-translational modifications (PTMs), such as phosphorylation, cleavage, methylation, ubiquitination, or acetylation as a function of cellular signaling activities in normal or pathological biological processes. PTMs may activate or inactivate signaling or other functions of the protein. As their name suggests, modification-specific antibodies will react with a target that carries a specific type of modification at a specific amino acid residue, without reacting with the unmodified target. It can be useful to design experiments incorporating both the modification-specific antibody and another antibody that recognizes total (modified + unmodified) expression levels of the target protein in order to confirm the change in readout of the modified antibody is the result of changes in activation and not changes in target expression (Figure 3).
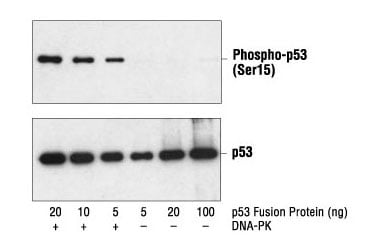 Figure 3. Western blot analysis of a p53 fusion protein, untreated or phosphorylated by DNA-PK, using Phospho-p53 (Ser15) Antibody #9284 (upper) and p53 Antibody #9282 (lower).
Figure 3. Western blot analysis of a p53 fusion protein, untreated or phosphorylated by DNA-PK, using Phospho-p53 (Ser15) Antibody #9284 (upper) and p53 Antibody #9282 (lower).
Other antibodies have been developed with specificity against point mutations and other types of mutations. Point mutation-specific antibodies will only react with target proteins bearing a specific amino acid substitution. Fusion-specific antibodies that only recognize proteins resulting from in-frame fusion mutations, such as the SS18-SSX fusion protein, are less common. Truncation mutations could be probed by designing experiments that incorporate antibodies that recognize the N- vs C-terminus of the truncated protein. Antibodies that recognize mutations involved in disease processes such as cancers may go on to be developed as clinical diagnostic tools (see below).
Species Reactivity
Not to be confused with antibody host species, which is described in the next paragraph, species reactivity refers to the species of the target protein. For example, many proteins expressed in mice will have homologs, or evolutionarily related proteins, expressed in other mammalian species. In some cases, an antibody raised against the mouse antigen will only react with the mouse homolog and not human or rat homologs of that protein. Other antibodies will be able to react with the homologs from multiple species. Whether an antibody has broad or narrow species reactivity is influenced primarily by the degree of sequence conservation in the epitope; a highly conserved epitope is more likely to result in an antibody that can be validated for the target in multiple species.
You will want to select an antibody with the species reactivity appropriate for your experimental model. So, for experiments with mouse cell lines, you would look for mouse species reactivity but if using human cell lines, you’d look for human reactivity. In the case of an exogenous expression model, such as expressing a human protein in a mouse cell line, you would need to select an antibody that has human reactivity and also does not react to the mouse homolog, although alternative experimental designs such as knockout of the mouse homolog or engineering of an epitope tag into the exogenously expressed protein are also possible.
Antibody Host Species
Antibodies can also be classified according to the species that produced them. Rabbit, mouse, and rat are the host species most commonly used to generate antibodies for research, especially in the case of monoclonal antibodies. This is partly because animal husbandry for these species is relatively straightforward. Although polyclonals may also be produced in mice or rats, they are typically made from larger animals such as rabbits, goats, donkeys, or sheep, from which it is easier to collect large volumes of sera.
A common approach to multiplexed immunoassays combines primary antibodies for distinct targets, each from a different host species, with secondary antibodies that are conjugated to distinct fluorophores. In this experimental design, each of the secondary antibodies will only recognize one of the primary antibodies, based on the species specificity of the constant region in the primary antibody.
For example, a "goat anti-mouse IgG" secondary antibody is produced in a goat hos, and will recognize the primary antibody produced from a mouse host. Conjugation to fluorescent dyes enables the detection and readout of each target in a microscope, flow cytometer, or other instruments. It is vital, however, to keep track of key details such as host species and isotype, and all of the primary and secondary antibodies must be well-validated for their intended use. Otherwise, misleading results will be obtained.
Antibody Isotypes & Subclasses
In mammals, antibodies are classified into five main isotypes or classes according to the heavy chain they contain. These are known as IgG, IgA, IgD, IgE, and IgM, where the heavy chain is gamma, alpha, delta, epsilon, or mu respectively. The heavy chains differ in terms of the number and sequence of constant domains and the structure of the hinge region, and in some species allow for each antibody isotype to be further divided into sub-types (Figure 4).
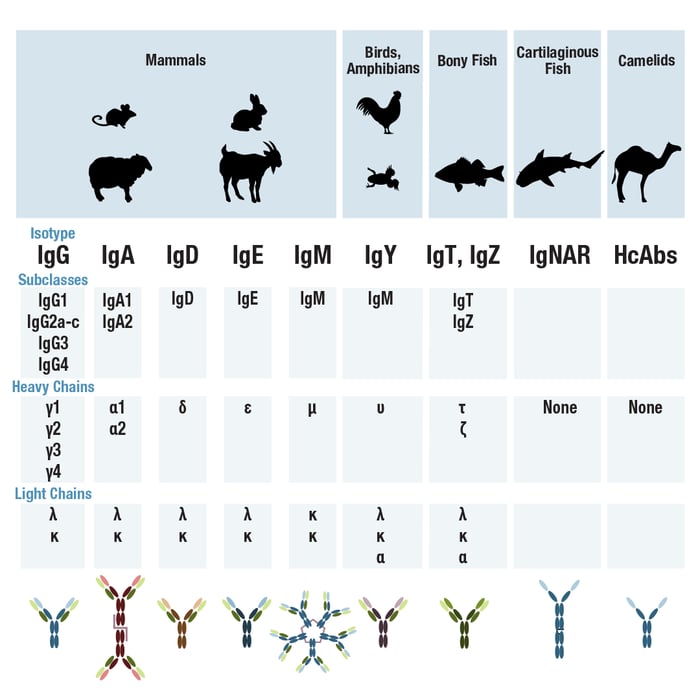
For example, in humans, the IgG isotype is grouped into four sub-types or subclasses—IgG1 through IgG4—whereas in mice, five different IgG sub-types exist. Rabbits have only one isotype: IgG. Antibodies with different target specificity, but the same sub-type, will have identical sequences in their constant domains. Consequently, they will produce the same Fc fragment if cleaved by pepsin. It's also possible for two or more antibodies from different species, isotypes, or subtypes to be generated against the same antigen.
By combining antibodies of different isotypes or subtypes within the same experiment, it is possible to add another layer of complexity to a multiplexed experimental design. For example, subclass-specific secondary antibodies that can distinguish between mouse IgG1 vs mouse IgG2a would be combined with primary antibodies from those subclasses.
It’s worth mentioning that in all mammals, the antibody light chains are classified as either kappa or lambda, based on the light chain gene locus selected during B cell maturation. The type of light chain is yet another antibody property that can be exploited to enable multiplexing. It’s also important to note that non-mammalian species have different antibody isotypes than mammals. For example, birds produce IgY antibodies, and bony fish, such as rainbow trout, produce IgT. Again, detecting the different isotypes relies on the availability of suitable, thoroughly validated secondary antibody reagents.
Finally, another major difference between antibody isotypes is the antibody avidity or antibody valency, i.e., the number of available antigen-binding sites. For example, while IgG, IgD, and IgE are bivalent, IgA has a valency of 4 and IgM has a valency of 10. As we saw in the first Antibody Essentials blog, and as illustrated again in Figure 5, the greater the valency, the more antigen an immunoglobulin molecule can bind, and the higher its avidity.
Clonality: Polyclonal vs Monoclonal Antibodies
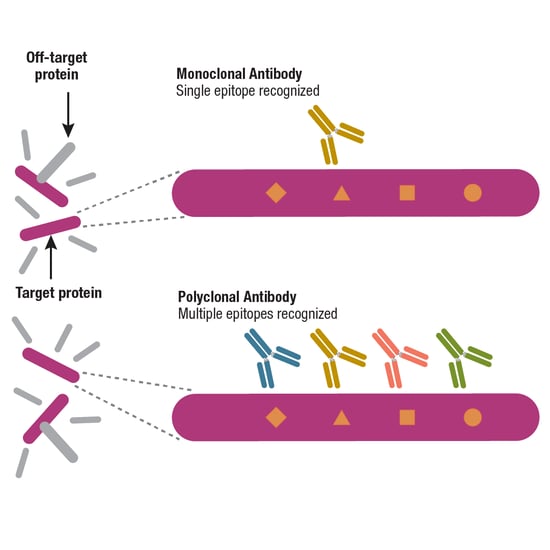
Another way of classifying antibodies considers whether they are polyclonal or monoclonal; the main distinction between the two refers to the B cells that generated them. Polyclonal antibodies are derived from mixed, or heterogeneous, B cell populations, meaning that a tube of polyclonal antibodies will actually contain many different antibodies. If the polyclonal is highly selective, the antibodies in this mixture will bind to multiple epitopes within the target protein, but not any other epitopes (Figure 5).
Monoclonal antibodies, on the other hand, are produced by identical B cells, so a tube of purified monoclonal antibody will contain multiple copies of the same antibody and should bind to a single epitope. The bottom line is that, when available, monoclonals offer more consistent performance than polyclonals, a topic that is explored in more detail in Part 4 of the Antibody Essentials series.
Recombinant Antibodies
One approach for further classifying monoclonal antibodies is to consider whether they were produced using conventional or recombinant methods. To be more precise, let’s first consider the difference between recombinant monoclonal antibody generation and production.
You may have read about the use of phage and yeast display libraries as an animal-free setting for high-throughput antibody discovery. This can be considered recombinant antibody generation. Note that recombinant monoclonal antibody production is not exclusively limited to antibodies that have been generated in vitro. A recombinant antibody could also be produced starting with the conventional immunization of an animal to evoke an immune response and expansion of B cells. Following isolation and clonal expansion of mature B cells (covered in Part 4 of Antibody Essentials), the genes for the heavy and light chains are cloned and sequenced. This is the distinguishing feature of a recombinant antibody: its gene and peptide sequences are known to the scientist developing it, enabling expression of the antibody in cell culture for scaled-up manufacturing. Often, the preferred choice for many research applications, recombinant monoclonal antibody production offers a continuous supply of an antibody, and more importantly for you, the end-user, they have greater lot-to-lot consistency and reliable performance.
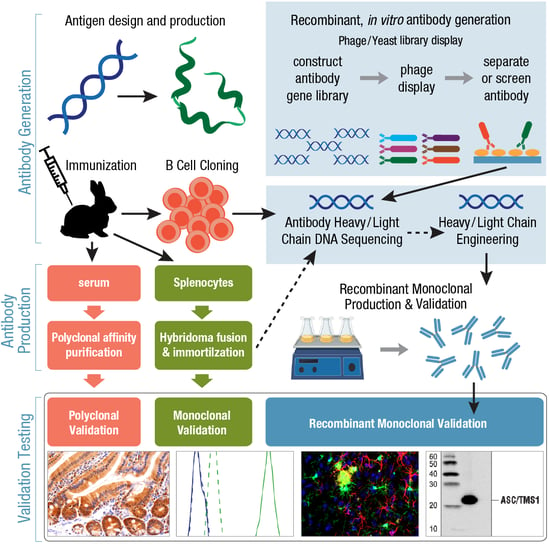
Since they are sequenced and amenable to engineering, the use of recombinant antibodies is seeing increased use as therapeutics, with a primary example being the development of antibody-drug conjugates. Other therapeutic approaches include species switching or humanizing antibodies developed in animal host species to reduce immunogenicity and the human anti-mouse antibody (HAMA) response; the production of single-chain variable fragments to maintain specificity while significantly reducing antibody size; and the generation of bispecific antibodies able to bind two different epitopes at once.
Research Use Only (RUO) Antibodies
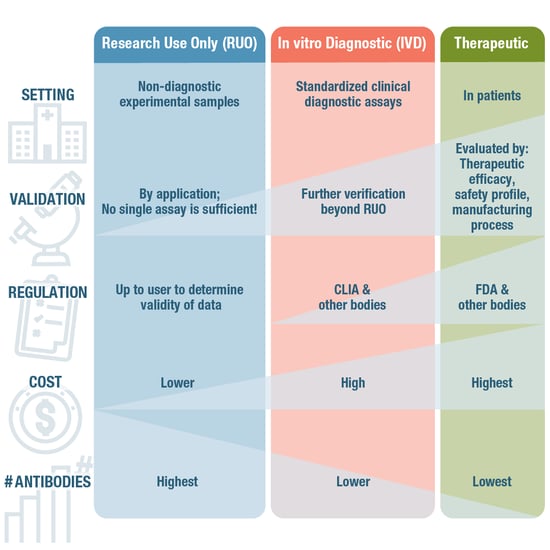
An important antibody classification concerns the setting in which the antibody will be used. If you are working in an academic research lab, you're most likely using antibodies labeled “research use only,” sometimes abbreviated as RUO. As opposed to therapeutic antibodies, which are covered below, ROU antibodies are intended for non-diagnostic, laboratory-based experiments (Figure 6, below).
Validation tests of RUO antibodies are designed according to the biological role of the target and use methods, protocols, and control materials that are relevant for the intended application. For instance, an RUO antibody deemed valid for western blotting should be supplied with clear experimental proof of its suitability for the western blot application, with data showing its performance against both positive and negative controls. It is important to note that validation in one application does not mean an antibody will necessarily work in other applications. For example, western blot validation data does not guarantee that an antibody will work for immunohistochemistry (IHC) or flow cytometry (IF). Therefore, it's critical that you look for validation data that are relevant to the application in which you will be using the antibody.
Further Reading: Learn more about validation techniques and the Hallmarks of Antibody Validation
Compared to therapeutic antibodies, RUO antibodies are generally not as stringently regulated, so it's ultimately up to the researcher to decide if the antibody validation data available from the manufacturer is sufficient. Improperly validated antibodies have contributed to the Antibody Reproducibility Crisis, so it's important to determine if you trust the antibodies used—and thus your experimental data—enough to publish the results.
Therapeutic Antibodies
In contrast to RUO antibodies, therapeutic antibodies are intended for use in patients. Therapeutic antibodies are special in that they have a biological activity that can be leveraged to treat disease. Mechanisms of action of a therapeutic antibody could be blocking ligand-receptor interactions, interrupting signaling cascades, or targeting tumor cells with radiation or different forms of cytotoxicity to kill them. This means that they require a very different approach to validation, or verification, as it’s more commonly referred to in this space. Rather than relying solely on application-specific validation, therapeutic antibodies are evaluated by their therapeutic efficacy, safety profile, and manufacturing process, whereby formal process characterization is used to assure patient safety in clinical trials.
In the US, this is tightly regulated by bodies such as the Food and Drug Administration (FDA), which has the authority to approve or reject a therapeutic antibody just like any other drug. As you can imagine, the cost of developing an FDA-approved therapeutic is much higher than an RUO antibody!
Clinical Diagnostic Antibodies
Positioned somewhere between RUO antibodies and therapeutic antibodies, diagnostic antibodies are used in a clinical setting on patient-derived samples. Here, they are an essential tool in standardized assays for the diagnosis of pathogenic infections, cancer, metabolic or immunological disorders, and many other medical conditions. Although some immunoassays, including IHC and ELISA, can incorporate either RUO or diagnostic use antibodies, clinical diagnostic use requires additional levels of verification and certification by a governing body such as Clinical Laboratory Improvement Amendments (CLIA) in the US.
Summary
Ultimately, the classification of an antibody is dependent on its method(s) of production, its intrinsic properties, and the applications it is suited to. All of these factors should be considered when choosing the best antibodies for your assays, and to meet your research and publication goals.
Coming next, we will be diving into the history and evolution of antibody technologies, and how it informs your decisions about antibody usage today in Antibody Essentials Part 3: How Antibody Technologies Evolved.
Read the entire Antibody Essentials series:
- Antibody Essentials Part 1: Antibody Basics
- Antibody Essentials Part 2: Antibody Diversity and Classification
- Antibody Essentials Part 3: How Antibody Technologies Evolved
- Antibody Essentials Part 4: Polyclonal vs Monoclonal Antibodies
- Antibody Essentials Part 5: Important Considerations for Antibody Selection
20-FLE-62535


